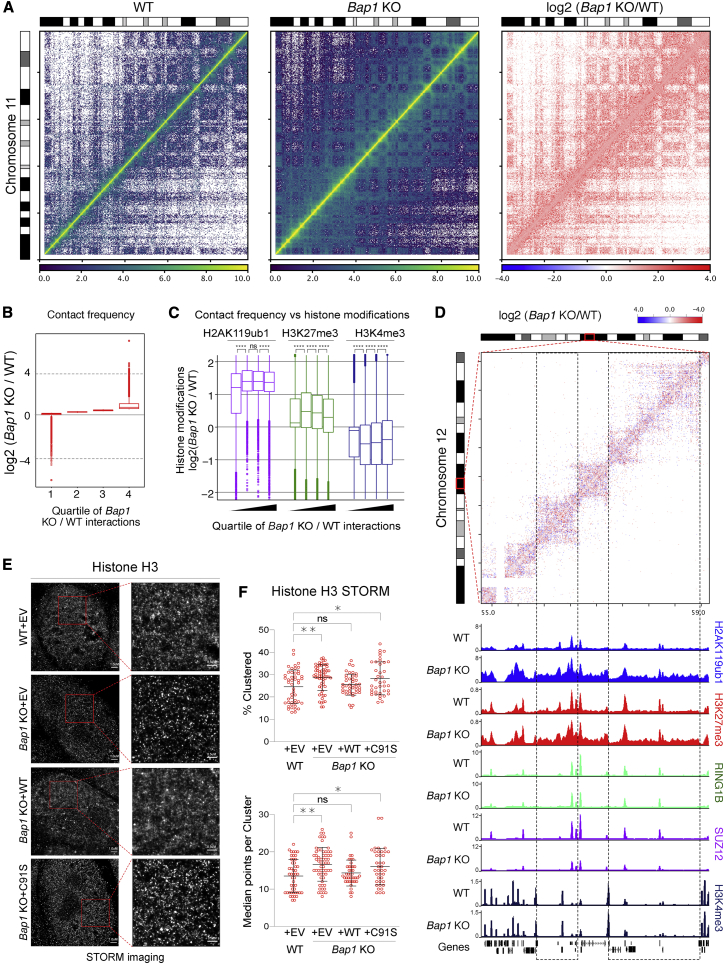
Figure 6.
Diffuse H2AK119ub1 accumulation causes global chromatin compaction
(A) Ice-normalized HiC contact matrix of the entire chromosome 11 in WT, Bap1 KO, and log2 fold change (BAP1 KO/WT) at 250 kbp resolution using two pooled HiC replicates.
(B) Boxplot of contact frequency of log2 fold change (Bap1 KO/WT) ratios divided into quartiles using two pooled HiC replicates.
(C) Boxplot of the log2 fold change (Bap1 KO/WT) ratio of the indicated histone modifications within the quartiles defined in (B). Wilcoxon test was used to ascertain significance.
(D) Top: Log2 fold change (Bap1 KO/WT) HiC contact matrix of the indicated region of chromosome 12 (54.8–59.15 Mb) at 10 kb resolution. Bottom: Genome browser snapshot of indicated ChIP-seq tracks at the same region of chromosome 12.
(E) Representative stochastic optical reconstruction microscopy (STORM) images of the indicated cell lines stained with histone 3. Scale bars of 1 μm (left column) and 0.5 μm (right column) are shown.
(F) STORM quantifications of percentage clustered (top) and median points per cluster (bottom) of H3. Data are represented as mean ± SD.
See also Figure S5.