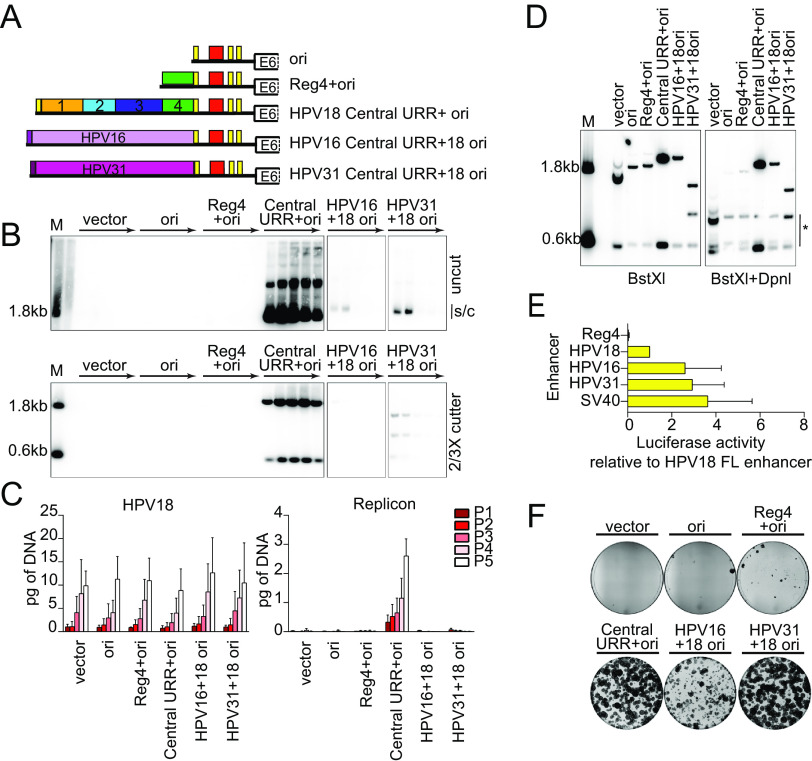
FIG 5.
The central URR from HPV16 and HPV31 does not fully support HPV18 replicon maintenance. (A) Diagram of enhancer replicons. (B) DNA collected from five passages of cotransfected cells were analyzed by Southern blotting as described in Fig. 2C. The monomeric, supercoiled form (s/c) of the replicon is indicated. (C) HPV18 genome (left) and replicon (right) copy numbers were measured by qPCR. (D) DNA was collected from keratinocytes cotransfected with HPV18 and the indicated test replicon 48 h after transfection. DNA samples were digested with BstXI or BstXI and DpnI and visualized by Southern blotting as described in Fig. 2C. DpnI digestion products are indicated by an asterisk. (E) Keratinocytes were transfected with luciferase reporter plasmids containing the cis elements indicated as well as a positive control (SV40 enhancer) and a negative control (empty luciferase vector containing a minimal promoter). Cell lysates were collected 48 h after transfection, and luciferase activity was measured and normalized to the total protein concentrations for each cell lysate and the negative-control plasmid. Reporter activity for each enhancer is shown relative to the HPV18 full-length enhancer. The luciferase assays shown were conducted at the same time as those reported in Fig. 4C, and the activity of the positive control and HPV18 enhancer luciferase results are replicated here. (F) Keratinocyte colonies arising from continuous G418 selection were stained with methylene blue approximately 14 days posttransfection. Error bars represent the standard deviation. The data shown are representative (panels B, D, and F) or an average of two (panel C) biological replicates. The average of three biological replicates is shown in panel E.