FIG 8.
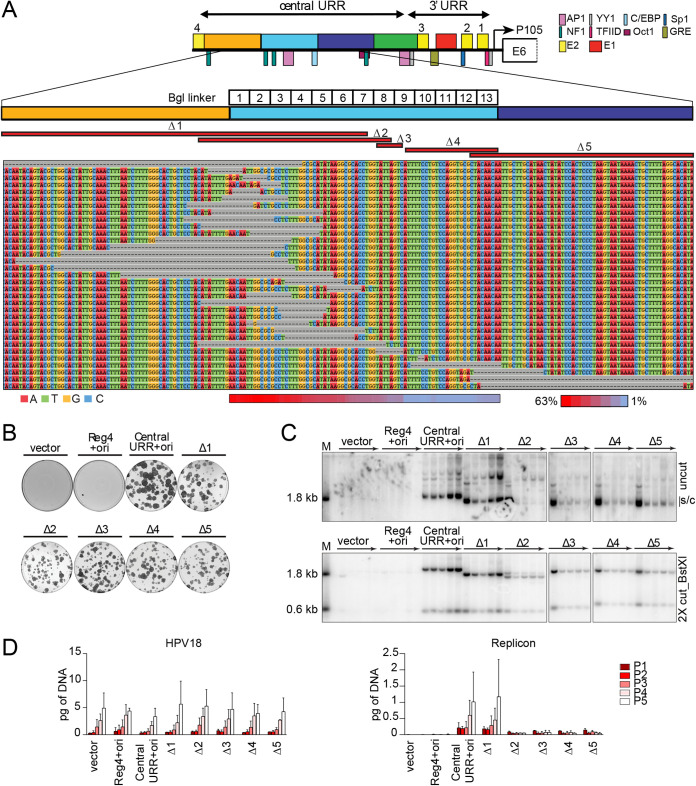
No single sequence in region 2 is absolutely required for replicon maintenance, but this region modulates replicon copy number and/or partitioning efficiency. (A) Diagram of the HPV18 central URR and ori in addition to the location of important viral (E1, E2) and cellular transcription factor binding sites. The locations of Bgl linker scanning mutations introduced into HPV18 region 2 are indicated by individually numbered white boxes (Bgl 1 to 13). A representative sequence alignment of deletions present in Bgl linker replicons recovered from passaged cells is shown. Deletions are highlighted by gray dashes, while nucleotides are individually colored as indicated. Individual deletions present in select replicons recovered from the Bgl chewback assay are mapped by red bars (labeled Δ1 through Δ5) above the sequence alignment. The frequency with which a region 2 nucleotide was deleted in the recovered replicons is indicated by a heatmap ranging from a maximum 63% (red) to 1% (blue) of the clones showing a deletion at a nucleotide site. (B) Keratinocyte colonies arising from continuous G418 selection were stained with methylene blue approximately 14 days posttransfection. (C) DNA collected from five passages of transfected cells were analyzed by Southern blotting as described in Fig. 2C. The monomeric, supercoiled form (s/c) of the replicon is indicated. (D) HPV18 genome (left) and replicon (right) copy numbers were measured by qPCR. Error bars represent the standard deviation. The data shown is representative (panels B and C) or an average (panel D) of two biological replicates.