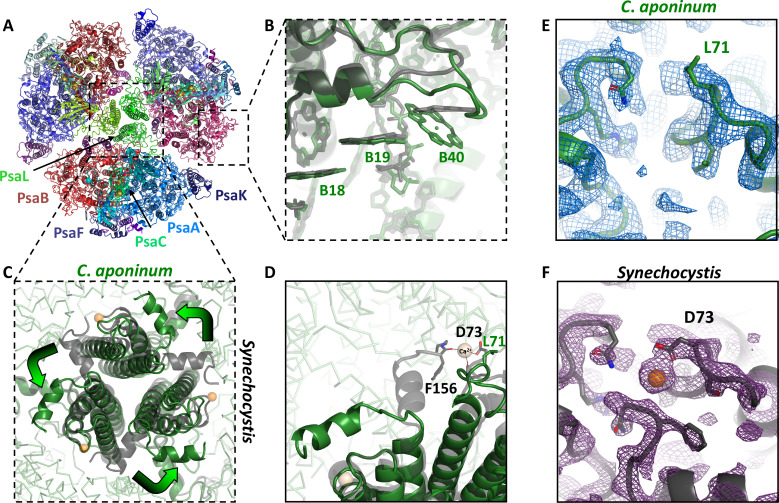
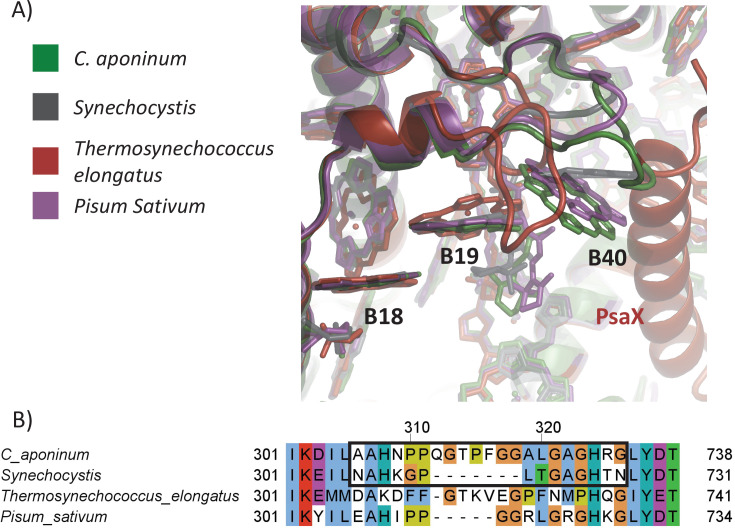
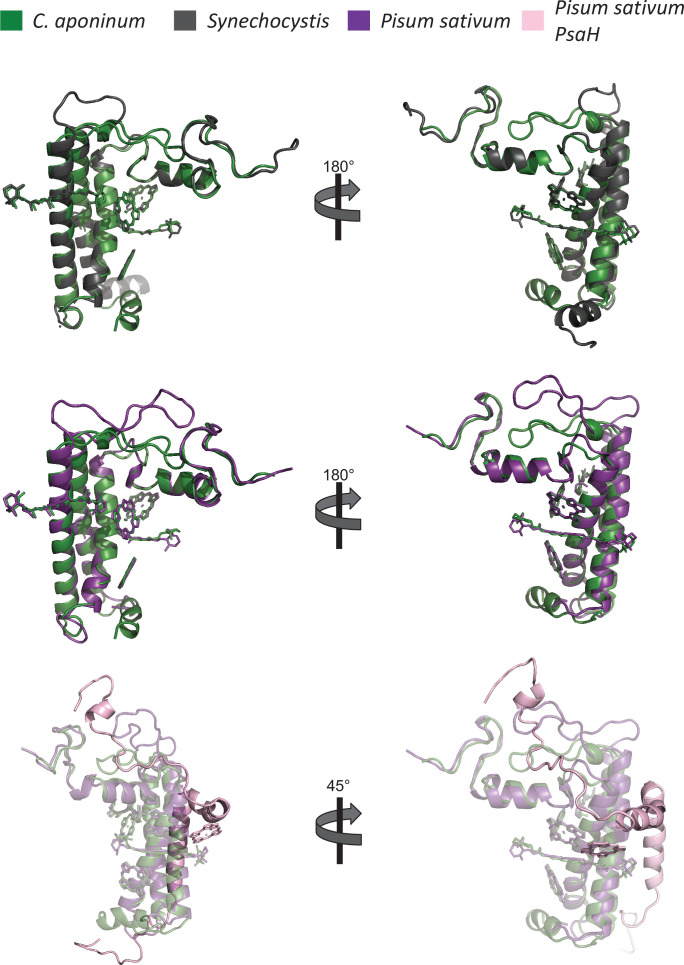
Figure 3. The structure of trimeric PSI from C. aponinum.
(A) C. aponinum trimeric PSI (B) chlorophyll B40 shifts its position due to the insertion seen in the PsaB subunit in C. aponinum (green) compared to Synechocystis (black). (C) The PsaL subunits of C. aponinum (green) and Synechocystis (black) showing the difference of the overall structure of the PsaL C-terminus. (D) The C-terminus of the PsaL subunit of C. aponinum (green) and Synechocystis (black) displaying the coordination to the Ca2+ in the adjacent monomer in Synechocystis, but is absent in C. aponinum and the Red_d mutant of Synechocystis. (E) C. aponinum and its electron density map compared to (F) Synechocystis (PDBID 5OY0, shown with 2Fo-Fc map) clearly depicting no density for the Ca2+ ion in the map for C. aponinum.