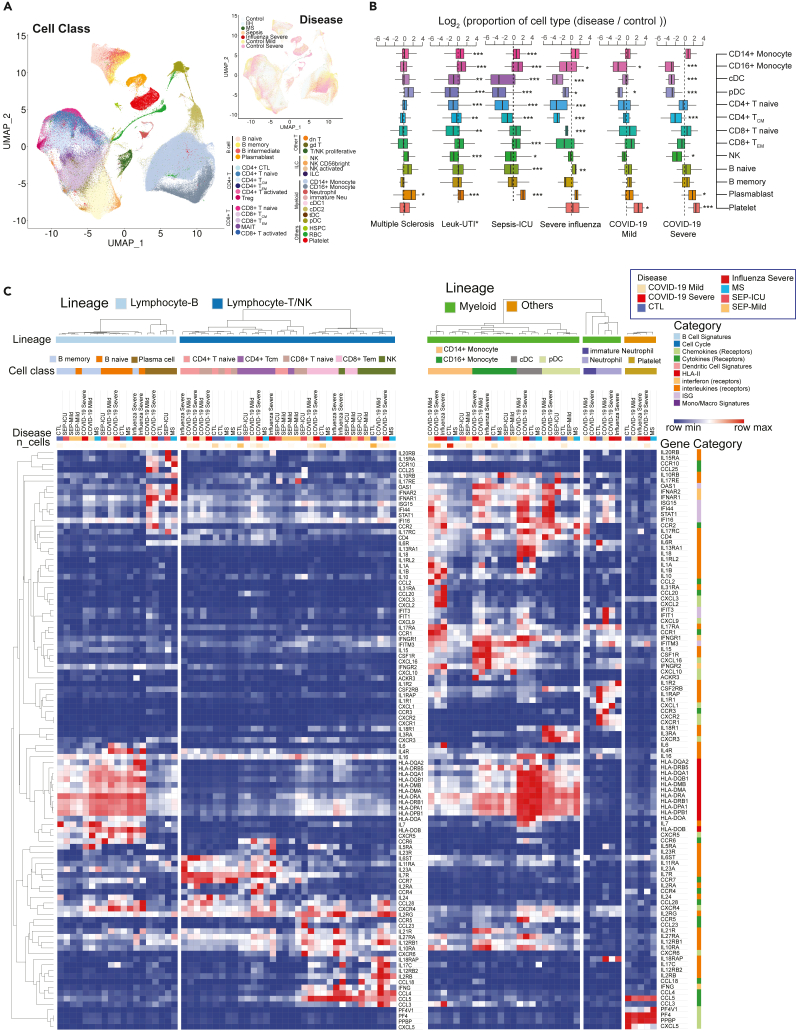
Figure 7.
Comparative analysis of differentially-expressed immunoregulatory genes between COVID-19 and other immune-mediated diseases
(A) Uniform Manifold Approximation and Projection (UMAP) shows the distributions of cell types (Left) and diseases (Top right) after the integration of datasets in multiple studies. MS: multiple sclerosis; IIH: idiopathic intracranial hypertension. IIH patients were recruited as controls in the multiple sclerosis study.
(B) Dynamic changes of immune cell types in different immune-mediated diseases compared to healthy controls. Log2(ratio) was calculated to show the levels of changes. ∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001. Statistical models can be found in the STAR Methods. Leuk-UTI: sepsis patients that enrolled into UTI with leukocytosis (blood WBC ≥12,000 per mm3) but no organ dysfunction. Box features are same with Figure 2B.
(C) Normalized expression values of key genes involved in immune signaling and responses are shown for cell types across multiple diseases. Lowly expressed genes (maximal average expression level across all cell types in the heatmap is less than 0.5 after Log2CPM normalization) were removed. See also Figure S18 and Table S7.