Abstract
In this narrative review, we present the hypothesis that key mutations in two genes, occurring 15 and 10 million years ago, were individually and then collectively adaptive for ancestral humans during periods of starvation, but are maladaptive in modern civilization (i.e., “thrifty genes”), with the consequence that these genes not only increase our risk today for obesity, but also for alcoholism. Both mutations occurred when ancestral apes were experiencing loss of fruit availability during periods of profound climate change or environmental upheaval. The silencing of uricase (urate oxidase) activity 15 million years ago enhanced survival by increasing the ability for fructose present in dwindling fruit to be stored as fat, which was a consequence of enhanced uric acid production during fructose metabolism that stimulated lipogenesis and blocked fatty acid oxidation. Likewise, a mutation in class IV alcohol dehydrogenase (ADH4) ~10 million years ago resulted in a remarkable 40-fold increase in the capacity to oxidize ethanol, which allowed our ancestors to ingest fallen, fermenting fruit. In turn, the ethanol ingested could activate aldose reductase that stimulates the conversion of glucose to fructose, while uric acid produced during ethanol metabolism could further enhance fructose production and metabolism. By aiding survival, these mutations would have allowed our ancestors to generate more fat, primarily from fructose, to survive changing habitats due to the Middle Miocene disruption and also during the late-Miocene aridification of East Africa. Unfortunately, the enhanced ability to metabolize and utilize ethanol may now be acting to increase our risk for alcoholism, which may be yet another consequence of once-adaptive thrifty genes.
Keywords: Alcoholism, Thrifty Genes, Fructose, Uric Acid, Uricase, Alcohol Dehydrogenase Class IV (ADH4)
Introduction
Nearly 60 years ago, the geneticist James Neel suggested that during periods of food shortage in our past, we may have acquired genes that improved our ability to store fat (i.e., thrifty genes), which would have aided survival at the time, but in today’s world may increase our risk for diabetes and obesity (Neel, 1962). Here, we suggest two genetic mutations in our past, causing a loss of uricase (urate oxidase) activity 15 million years ago (MYA) and a mutated class IV alcohol dehydrogenase (ADH4) having a 40-fold increase in the capacity for ethanol oxidation 10 MYA, may have acted as thrifty genes that not only increase our risk for obesity today, but that may also have a role in alcoholism.
The questions of when and how humans developed a preference for alcohol have historically become subjects of controversy. In the early 1950s, for example, American botanist Jonathan Sauer first challenged the longstanding assumption that bread was the impetus for agriculture by proposing that beer was instead the initial motivator (Braidwood et al., 1953). Recent archeological evidence suggests that beer was being made from wheat or barley as early as 13,000 years BP by the Natufian people prior to the introduction of agriculture (Liu et al., 2018), and possibly by others (based on evidence at a Neolithic temple sanctuary in Turkey) dating between 12,000 and 10,000 years BP (Dieterich et al., 2012). Wine and beer were also being made by early agricultural communities in Israel along the Black Sea, and in China, during the Neolithic period (McGovern et al., 2017, McGovern et al., 2004). This evidence supports the idea that a human preference for alcohol developed at least as early as the onset of agriculture, if not before it.
Additionally, alcohol may have been of evolutionary importance to our primate ancestors (Dudley, 2000). Ethanol from fermenting fruit, for example, would have provided both sensory cues and dietary calories (approximately 7 calories/g when assuming energy efficient metabolic pathways) (Pirola and Lieber, 1972) that could have aided our primarily frugivorous ancestors in routine foraging and during periods of food shortage (Dudley, 2000, Dudley, 2002, Dudley, 2004, Dudley, 2014, Dudley, 2020). However, most primates are limited as to how much ethanol they can ingest, given their metabolism. Multiple pathways are involved in metabolizing ethanol (Cederbaum, 2012), and many begin with one of several classes of alcohol dehydrogenase (ADH) that tend to metabolize different alcohols with varying affinities (Duester et al., 1999). For example, an ancestral version of ADH4 in our human lineage was very efficient at metabolizing geraniol, an alcohol produced by plants as an antifeedant in leaves, but inefficient at metabolizing ethanol (Carrigan et al., 2015). This changed around 10 MYA when the ancestor to humans and the African great apes evolved a mutation in ADH4 that was able to oxidize ethanol 40-fold better than before (Carrigan et al., 2015).
This ADH4 mutation is consistent with exposure to fermented carbohydrates, and ethanol may have been consumed by our primate precursors since as early as the middle to late Miocene. Here, we will review this unique evolutionary period and present the hypothesis that our ability to metabolize ethanol enabled our ancestors to obtain necessary calories from scarce fruit supplies during times of climate change and environmental upheaval. We support this concept by considering an additional mutation in the metabolic enzyme uricase that occurred during the middle Miocene, and that is involved in fruit metabolism (Johnson et al., 2020, Johnson and Andrews, 2015, Johnson and Andrews, 2010, Johnson et al., 2008). Both genes represent changes in gene activity rather than the acquisition of genes, as Neel initially hypothesized (Neel, 1962). However, as we have previously suggested, the uricase mutation acted as a thrifty gene by improving our ability to generate fat primarily from fructose (Johnson and Andrews, 2010). Here, we suggest the ADH4 mutation further improved our ability to store fat primarily from fructose.
To begin with, the ADH4 mutation allowed apes in our lineage to ingest and metabolize more ethanol (Carrigan et al., 2015). Importantly, it has been recently shown that ethanol stimulates the production of fructose in the liver by activating aldose reductase that stimulates the conversion of glucose to fructose (Wang et al., 2020), which would provide another source of fructose as fruit became less available. Uric acid is also generated during ethanol metabolism and can provide feedback to increase endogenous fructose production (Sánchez Lozada et al., 2019) and stimulate fructose metabolism (Lanaspa et al., 2012c). Fructose and ethanol are also known to induce similar craving behavior, and increasing evidence suggests ethanol and fructose are physiologically intertwined (i.e., fructose is “alcohol without the buzz”) (Lustig, 2013). Thus, we present the hypothesis that, in addition to increasing our risk for diabetes and obesity, as Neel first suggested, the mutations in uricase and ADH4 also increased our risk for alcoholism.
Search Strategy
For this narrative review, PubMed and Google Scholar were first searched for articles containing combinations of terms relating to alcohol or ethanol, and thrifty genes. PubMed and Google Scholar were then searched for articles containing combinations of terms relating to uricase/urate oxidase and ADH4. PubMed and Google Scholar were searched a third time for articles containing combinations of terms relating to alcohol/ethanol and fructose, including alcohol/ethanol and fructose with or without uric acid. Articles, and reference lists within articles, were evaluated for relevance to our topic, nonsystematically.
Results and Discussion
Based on our review of the literature, few studies consider whether thrifty genes may increase our risk for alcoholism (You and Arteel, 2019, Nunn et al., 2009, Ehlers and Wilhelmsen, 2007, Ferré and Foufelle, 2007). With the arguable exception of one study considered here (Carrigan et al., 2015), no study explicitly considers the role of thrifty genes with respect to alcohol and the origins of our hominid ancestors. Here, to our knowledge for the first time, we consider together two mutations, occurring during the middle and late Miocene, in uricase and ADH4, respectively. These mutations would have individually and then collectively helped our hominoid and hominid ancestors survive by increasing their abilities to obtain necessary calories from scarce supplies of fruit, including fermenting fruit. Unfortunately, the enhanced ability for our ancestors to metabolize and utilize the ethanol in fermenting fruit millions of years ago may be increasing our risk for alcoholism today.
Climate Change Leads to an Environmental Crisis in the Middle Miocene
The early Miocene, i.e., 24–18 MYA, represented a golden age in primate evolution. Apes (Hominoidea) first appeared during this time in East Africa, having diverged from a common anthropoid ancestor with the cercopithecine monkeys while evolving larger bodies, greater cranial capacities, and the absence of tails. These early apes lived in trees in tropical rainforests on a diet consisting primarily of fruit, and rapidly diversified to at least 14 known genera by 17 MYA (Begun, 2003). Around this same time (19–17 MYA), sea levels fell in association with global cooling and allowed numerous species, including apes, to migrate out of Africa into Eurasia (Andrews and Kelley, 2007). The immigrant apes were able to continue subsisting on predominately fruit-based diets in subtropical woodland environments that existed in Eurasia at the time (Andrews, 2015).
Global cooling from the early-Miocene high led to a climatic crisis (i.e., the Middle Miocene disruption, from 15–12 MYA) that resulted in extinction of up to 30% of all mammals (Andrews, 2015). For the apes living in Africa, falling temperatures resulted in a contraction of their ranges, but the climate overall was still sufficiently warm and wet to sustain fruit production through the year. As a consequence, evolutionary pressures among the African apes were relatively minimal in the center of their range (Johnson and Andrews, 2010).
In contrast, the fall in temperature in western Eurasia (and particularly in Europe) was more severe, resulting in greater climatic seasonality and a change in habitat toward seasonal deciduous forests with open grasslands (Andrews, 2015). Fruit availability was reduced, especially during the cooler months (possibly from the loss of fig trees, which can fruit all year long) (Johnson and Andrews, 2015). Many dental remains of these apes show evidence for intermittent (likely seasonal) starvation, as noted by linear bands of enamel hypoplasia on the incisors (Kelley, 2008, Skinner et al., 1995). The consequence was a retreat of different ape species to various refugia, followed by the eventual extinction of the apes in western Eurasia by about 8 MYA (Agusti et al., 2003). However, fossil evidence suggests that, while some ape taxa may have moved to Southeast Asia and evolved into the orangutan lineage, others may have emigrated from Eurasia back to Africa as predecessors of gorillas, chimpanzees, and humans (thus the “Back-to-Africa” hypothesis) (Andrews and Kelley, 2007, Begun, 2000). One potential candidate for the return to Africa is Kenyapithecus kizili, for which (and despite the generic name) the earliest fossil evidence is from Turkey, followed by an appearance in Africa two million years later (Kelley, 2008, Kelley et al., 2008, McCrossin and Benefit, 1997).
How Mutations in Fructose and Alcohol Metabolism May Have Aided Hominoid and Hominid Survival in the Middle Miocene Disruption and Late-Miocene Aridification of East Africa
The changing environment in western Eurasia was more severe than in Africa for hominoid apes during the middle Miocene. Changing forests and open grasslands affected fruit availability, especially during the cooler months of the year. As fruits became less available, the apes had to spend more time on the forest floor, where they knuckle-walked to find fallen fruit as well as other food sources such as tubers and roots (i.e., fallback foods) (Marshall and Wrangham, 2007). This may have led to changes in dentition observed during this period, possibly to accommodate harder foods (Andrews, 1990, King et al., 1999, Ersoy et al., 2008). Consistent with the fossil evidence, there were also multiple genetic mutations during the middle Miocene (Samonte and Eichler, 2002).
First Mutation: Uricase
One relevant mutation was in the uricase gene, which controls an enzyme that regulates serum and intracellular uric acid levels (Oda et al., 2002, Kratzer et al., 2014). The loss of a functioning version of this enzyme in our lineage began in the Oligocene as a series of mutations that affected the promoter, until the gene was completely silenced into a pseudogene around 15–12 MYA (Kratzer et al., 2014). The mutation affected the common ancestor of humans and all great apes (including the orangutan), whereas a similar mutation knocked out uricase in the ancestors of lesser apes (Oda et al., 2002). The effect of the uricase mutation was to double serum uric acid levels in apes, relative to other mammals, from 1 to 2 mg/dL to approximately 3 to 4 mg/dL (Johnson et al., 2005). Indeed, studies of Yanomami hunter-horticulturalists living on a native diet of plantains, other fruits, and wild game show serum uric acid levels around 3 mg/dL, similar to what is observed in great apes (Johnson et al., 2005). However, the loss of uricase has made it more difficult to control serum uric acid levels in modern civilizations. In particular, the Western diet is high in purines, sugar, and alcohol, all three of which can raise serum uric acid (Johnson et al., 2005). As such, serum uric acid levels are currently much higher in industrialized populations, in the 4 to 6 mg/dL range, with more than 20 million people showing serum uric acid levels of 7 mg/dL (Chen-Xu et al., 2019).
The observation that uricase mutations occurred in both the human/great ape and lesser ape lineages suggested that the loss of uricase must have provided an evolutionary advantage. While various hypotheses have been proposed (Orowan, 1955, Ames et al., 1981), recent studies suggest that knocking out the uricase gene may have enhanced metabolic capacity of the apes in response to decreasing fruit availability. A major nutrient in fruit is fructose, which is distinct from most nutrients in that it triggers an adenine nucleotide turnover reaction that leads to the generation of uric acid. Specifically, the rapid consumption of adenosine triphosphate (ATP) in the first step of fructose metabolism leads to local phosphate depletion, which causes the rapid removal of adenosine monophosphate (AMP) to generate uric acid (Figure 1) (Van den Berghe, 1986, Maenpaa et al., 1968). The removal of AMP causes persistent local ATP depletion that is amplified by uric acid, which causes mitochondrial oxidative stress and inhibition of AMP-activated protein kinase, both which would act to reduce the ability to replenish intracellular ATP levels (Lanaspa et al., 2012a, Lanaspa et al., 2012b, Cicerchi et al., 2014). Consequences of the adenine nucleotide turnover and mitochondrial oxidative stress induced by fructose include the stimulation of lipogenesis (Lanaspa et al., 2012b, Softic et al., 2018), an impairment in fat oxidation (Lanaspa et al., 2012c, Softic et al., 2019), and the development of insulin resistance (Softic et al., 2018, Softic et al., 2019).
Figure 1. Fructose Metabolism Generates Uric Acid.
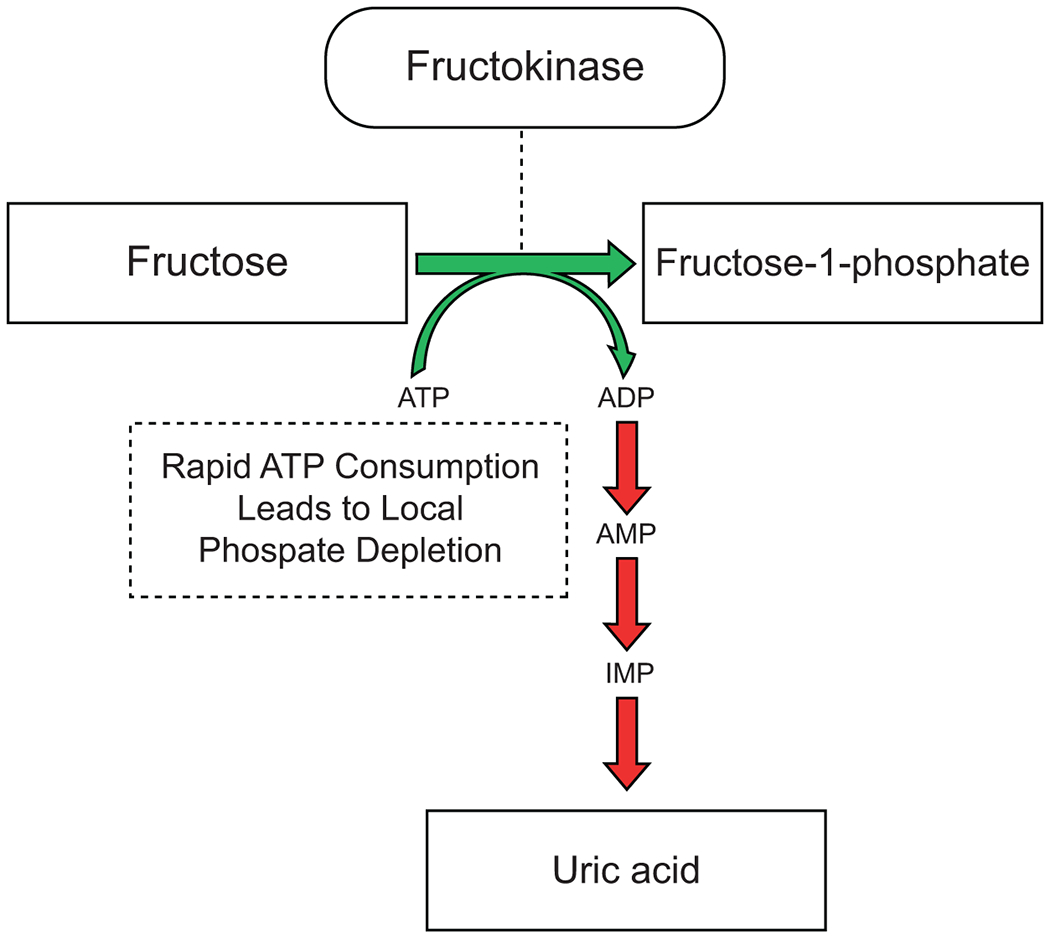
The rapid consumption of ATP in the first step of fructose metabolism leads to local phosphate depletion, which then causes rapid removal of AMP to generate uric acid.
Inhibition of uricase results in an amplification of this uric acid response (Stavric et al., 1976), which heightens the ability of fructose to induce metabolic syndrome, for example, in the uricase-inhibited rat (Tapia et al., 2013). We also resurrected the original uricase gene to study its function in human liver cells and found that, while there was still an increase in fat in response to fructose, the amount of fat was doubled by the same amount of fructose in the gene’s absence (Kratzer et al., 2014). Absence of the ancestral uricase gene also resulted in enhanced gluconeogenesis and insulin resistance (Cicerchi et al., 2014) that likely provided a survival advantage by preferentially shunting glucose away from insulin-dependent muscles to the brain, as most regions of the brain do not require insulin for glucose uptake (Sprengell et al., 2021, Gray et al., 2014, Seaquist et al., 2001, Hasselbalch et al., 1999).
These studies suggest that the uricase mutation likely provided a survival advantage by allowing ancestral hominoids to make the most fat possible from declining available fruits. However, these particular ancestors would have also been constrained in their digestion of fermenting fruits, given the presence of ADH forms that were relatively inefficient at metabolizing ethanol.
Second Mutation: ADH4
When fruit ripens, intense ecological competition begins for the accessible simple sugars within the pulp. This is true, of course, for the animals that primarily eat fruit (including many primates) and fulfill the evolutionary objective of seed dispersal for the plant. It is also true for microbes, among which yeasts—especially Saccharomyces cerevisiae and its close relatives—are the dominant group (Dashko et al., 2014, Piskur et al., 2006).
At least three lineages of yeasts—including the Saccharomyces cerevisiae lineage—appear to have independently evolved strong reductions in their oxidative respiratory capacities (i.e., the Crabtree effect), resulting in the fermentation of pyruvate to ethanol even in the presence of oxygen (Hagman et al., 2013, Rhind et al., 2011, Rozpędowska et al., 2011, Piskur et al., 2006, Ihmels et al., 2005). These lineages also evolved greater tolerances to ethanol than other microbes (Casey and Ingledew, 1986, Ingram and Buttke, 1985). This outcome created an evolutionary advantage by which these yeasts can quickly convert sugars into ethanol to kill off their bacterial competition (Ingram and Buttke, 1985). When the sugars begin to run out, some of the yeasts are also able to convert the ethanol into energy (Pfeiffer and Morley, 2014, Dashko et al., 2014, Rozpędowska et al., 2011, Thomson et al., 2005, de Jong-Gubbels et al., 1996). The Saccharomyces yeasts, for example, evolved their ability to consume ethanol via a duplication of ADH that took place during the Cretaceous period when fleshy fruits arose (Thomson et al., 2005), as did the fermentative capacity of the Saccharomyces yeasts (Benner et al., 2002).
Concurrently in the Cretaceous, the first primates evolved (Steiper and Young, 2006). As with the yeasts, primates appear to have coevolved with the angiosperms that produce fleshy fruits (Sussman et al., 2013, Tiffney, 1984), and fruit consumption may have been an underpinning to the evolution of larger primate brains (DeCasien et al., 2017). However, primates mostly eat ripe fruits in trees, whereas fruits with the highest populations of yeasts and other microbes are mostly the overripe and usually damaged fruits on the ground (Carrigan et al., 2015, Dudley, 2004). There are still very few measurements of ethanol in the wild, but the Astrocaryum standleyanum palm in the lowland Panamanian rainforest, for example, exhibits pulp-ethanol concentrations of 0.6% for ripe fruit hanging in trees, 0.9% for ripe fallen fruits, and 4.5% for overripe fallen fruits (the highest was 8.1% in an overripe fallen fruit) (Dudley, 2004, Dudley, 2002).
For our primate ancestors residing mostly in the trees, the higher amounts of ethanol in fallen fruits would have rarely been encountered, and alcohol-metabolizing enzymes like ADH would have therefore been more useful for breaking down antifeedant alcohols produced by the plants themselves (Carrigan et al., 2015). As long as those enzymes were also inefficient at metabolizing ethanol, our ancestors would probably have needed to avoid overly alcoholic foods. Indeed, at least some frugivores (including primates in Southeast Asia lacking the ADH4 mutation of humans and the African great apes) tend to avoid fruits higher in ethanol (Dominy, 2004, Sánchez et al., 2004). Others, including at least one other primate (the aye-aye) that independently evolved the same ADH4 mutation, prefer to consume higher ethanol concentrations (Gochman et al., 2016). Another primate (the slow loris) and a closely related non-primate mammal (the pentailed treeshrew) have been observed regularly consuming large quantities of fermenting palm nectar while showing no signs of intoxication (in the case of the pentailed treeshrew, which consumed the most ethanol, it also produced alcoholic-level quantities of a secondary ethanol metabolite, ethyl glucuronide) (Wiens et al., 2008). Even our closest primate relatives, the chimpanzees, have been known to use leaves as cups to steal intentionally fermented palm sap from neighboring humans (Hockings et al., 2015). As for the fruit-eating common ancestors we share with the chimpanzees and gorillas, their ability to break down ethanol would have been most useful when consuming fallen fruit that has been fermenting for a longer time.
If the ancestors of gorillas, chimpanzees, and humans did indeed return to Africa from Eurasia, they would have passed through an East Africa much different from that at the end of the early Miocene (Figure 2) (Andrews, 2015). During the middle Miocene, East Africa began experiencing climatic change distinct from the rest of Africa due to volcanic activity along the East African Rift. Uplift, mountains, and large lakes along the rift contributed to gradual aridification. This caused a progressive increase in deciduous woodland habitats and bushland throughout East Africa during the middle to late Miocene, ultimately resulting in a savanna-dominated habitat within which our hominid ancestors evolved (Andrews, 2015, Begun, 2003, Johnson and Andrews, 2010). About 10 MYA, East Africa would have been in the middle of this transition (Johnson and Andrews, 2010, Carrigan et al., 2015). The common ancestor of African apes and humans would have experienced ever-increasing seasonality, in the process becoming less arboreal with greater access to fallen and fermenting fruit on the woodland floor (Carrigan et al., 2015, Andrews, 2015).
Figure 2. Apes Evolve Mutated Uricase and ADH4 Genes During the Miocene.
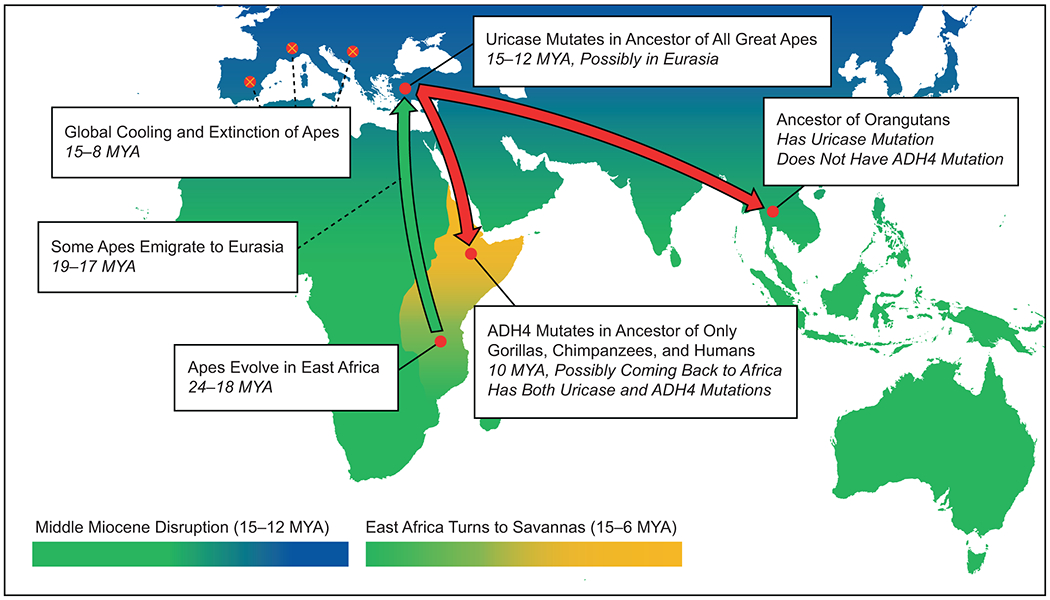
All the extant great apes (orangutans, gorillas, chimpanzees, and humans) exhibit the uricase mutation, which would have been advantageous during the Middle Miocene disruption in Europe. Only the African apes (gorillas, chimpanzees, and humans) also have the ADH4 mutation, which would have secondarily been useful during the late-Miocene aridification of East Africa.
It was at this time, around 10 MYA, that the mutation in ADH4 likely occurred, thereby increasing the capacity for ethanol oxidation 40-fold (Carrigan et al., 2015). Using a maximum-likelihood analysis, Carrigan et al. inferred the amino acid sequence of nine ancestral ADH4 enzymes, and then synthesized and examined each of them for their kinetic properties (Carrigan et al., 2015). The group found a single amino acid replacement (A294V) in the ADH4 of the last common ancestor of humans, chimpanzees, and gorillas, which dramatically increased catalytic activity toward ethanol (Carrigan et al., 2015). The increase is due primarily to a drop in the Michaelis–Menten constant for ethanol, from values exceeding 1,000 mM in all previous ancestral versions of ADH4, down to 43 mM (± 5.8 mM) in the version that mutated 10 MYA (Carrigan et al., 2015). The mutated ADH4 remains the predominant ADH4 in modern humans and gorillas, and is one of two common polymorphisms in chimpanzees (Carrigan et al., 2015). Unlike the uricase mutation, the ADH4 mutation is not carried by orangutans (Carrigan et al., 2015), which means it occurred after orangutans split from the other great apes, and possibly following a return to Africa.
The presence of the ADH4 mutation would have provided a survival advantage as it would have enhanced the ability of our hominid ancestors to benefit from fermenting fruit, thus improving foraging capacity and further selecting for terrestriality (Carrigan et al., 2015). In addition to effectively increasing our tolerance for the ethanol in fermenting fruit, however, we suggest that the mutated ADH4 would have further improved our ability to generate fat from fructose, both from the fructose in the fermenting fruit, and also, for example, from the endogenous fructose generated because of ethanol. As mentioned, it has recently been shown that ethanol stimulates the production of fructose in the liver by activating aldose reductase that stimulates the conversion of glucose to fructose, which would have led to increased fat production from fructose (Wang et al., 2020). Uric acid is also generated during ethanol metabolism (Yamamoto et al., 2005) and can provide feedback to increase endogenous fructose production (Sánchez Lozada et al., 2019) and stimulate fructose metabolism (Lanaspa et al., 2012c), causing even more fat generation. Finally, in addition to having similar metabolic effects, ethanol and fructose are known to similarly induce craving behavior (i.e., fructose is “alcohol without the buzz”) (Lustig, 2013). Ethanol and fructose are thus physiologically intertwined.
Conclusions and Future Directions
Uricase and ADH4 mutations may have been targets of selection for human ancestors that aided survival via improved metabolic capacity during times of profound climate change or environmental upheaval. Unfortunately, the enhanced ability to metabolize and utilize fructose and ethanol may now be acting to increase our risk for excessive consumption. Today, we no longer primarily consume the fructose or ethanol in wild fruits. Fructose is also present, for example, in table sugar (in the dimer form of sucrose), high-fructose corn syrup, and other added sugars, the intake of which has historically soared, resulting in increased risk for diseases like obesity and diabetes (Johnson et al., 2017). As for ethanol, the development of the ADH4 mutation has allowed many humans to consume large quantities of the ethanol in alcoholic beverages, with all of its varied consequences, including those involving fructose.
We recommend that future research test the roles of fructose and urate metabolism in alcohol-induced behavior and fat metabolism. We also recommend researching similar mutations that may have occurred in other enzymes involved in ethanol metabolism, including other classes of ADH enzymes, cytochrome P450 2E1 (CYP2E1) of the microsomal ethanol oxidizing system (MEOS), catalase, and the aldehyde dehydrogenase enzymes that break down the first oxidation product of ethanol metabolism, acetaldehyde (Teschke, 2019, Peana et al., 2017, Cederbaum, 2012).
In conclusion, we suggest ethanol is physiologically intertwined with fructose, a major nutrient from our frugivorous past that, along with ethanol, is causing addictive problems for many humans today. Thus, alcoholism may reflect yet another contemporary and adverse consequence of once-adaptive selection on thrifty genes.
Support:
This work was supported in part by National Institutes of Health 1U01AA027997-01 (RJJ, DRT, and MAL). The grant is supporting research on the relationship of sugar and alcohol metabolism.
Footnotes
Conflict of Interest: RJJ and MAL have equity with Colorado Research Partners, LLC that is developing fructose inhibitors, and RJJ also has equity with XORTX therapeutics that is developing novel xanthine oxidase inhibitors. RJJ has also consulted for Horizon Pharma. All other authors declare no conflicts of interest.
References
- Agusti J, Sanz de Siria A, Garces M (2003) Explaining the end of the hominoid experiment in Europe. J Hum Evol 45:145–153. [DOI] [PubMed] [Google Scholar]
- Ames BN, Cathcart R, Schwiers E, Hochstein P (1981) Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci U S A 78:6858–6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P (1990) Palaeoecology of the Miocene fauna from Paşalar, Turkey. Journal of Human Evolution 19:569–582. [Google Scholar]
- Andrews P (2015) An apes’s view of human evolution. Cambridge University Press, Cambridge. [Google Scholar]
- Andrews P, Kelley J (2007) Middle Miocene dispersals of apes. Folia Primatologica; International Journal of Primatology 78:328–343. [DOI] [PubMed] [Google Scholar]
- Begun DR (2000) Middle Miocene hominoid origins. Science 287:2375. [PubMed] [Google Scholar]
- Begun DR (2003) Planet of the apes. Sci Am 289:74–83. [DOI] [PubMed] [Google Scholar]
- Benner SA, Caraco MD, Thomson JM, Gaucher EA (2002) Planetary biology—paleontological, geological, and molecular histories of life. Science 296:864. [DOI] [PubMed] [Google Scholar]
- Braidwood RJ, Sauer JD, Helbaek H, Mangelsdorf PC, Cutler HC, Coon CS, Linton R, Steward J, Oppenheim AL (1953) Symposium: did man once live by beer alone? American Anthropologist 55:515–526. [Google Scholar]
- Carrigan MA, Uryasev O, Frye CB, Eckman BL, Myers CR, Hurley TD, Benner SA (2015) Hominids adapted to metabolize ethanol long before human-directed fermentation. Proc Natl Acad Sci U S A 112:458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey GP, Ingledew WM (1986) Ethanol tolerance in yeasts. Crit Rev Microbiol 13:219–280. [DOI] [PubMed] [Google Scholar]
- Cederbaum AI (2012) Alcohol metabolism. Clin Liver Dis 16:667–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Xu M, Yokose C, Rai SK, Pillinger MH, Choi HK (2019) Contemporary prevalence of gout and hyperuricemia in the United States and decadal trends: The National Health and Nutrition Examination Survey, 2007-2016. Arthritis Rheumatol 71:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicerchi C, Li N, Kratzer J, Garcia G, Roncal-Jimenez CA, Tanabe K, Hunter B, Rivard CJ, Sautin YY, Gaucher EA, Johnson RJ, Lanaspa MA (2014) Uric acid-dependent inhibition of AMP kinase induces hepatic glucose production in diabetes and starvation: evolutionary implications of the uricase loss in hominids. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 28:3339–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashko S, Zhou N, Compagno C, Piskur J (2014) Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res 14:826–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong-Gubbels P, van Dijken JP, Pronk JT (1996) Metabolic fluxes in chemostat cultures of Schizosaccharomyces pombe grown on mixtures of glucose and ethanol. Microbiology (Reading) 142 ( Pt 6):1399–1407. [DOI] [PubMed] [Google Scholar]
- DeCasien AR, Williams SA, Higham JP. Primate brain size is predicted by diet but not sociality. Nat Ecol Evol [serial online]. 1:112. [DOI] [PubMed] [Google Scholar]
- Dieterich O, Heun M, Notroff J, Schmidt K, Zarnkow M (2012) The role of cult and feasting in the emergence of Neolithic communities. New evidence from Göbekli Tepe, south-eastern Turkey. Antiquity 86:674–695. [Google Scholar]
- Dominy NJ (2004) Fruits, fingers, and fermentation: the sensory cues available to foraging primates. Integr Comp Biol 44:295–303. [DOI] [PubMed] [Google Scholar]
- Dudley R (2000) Evolutionary origins of human alcoholism in primate frugivory. Quarterly Review of Biology 75:3–15. [DOI] [PubMed] [Google Scholar]
- Dudley R (2002) Fermenting fruit and the historical ecology of ethanol ingestion: is alcoholism in modern humans an evolutionary hangover? Addiction 97:381–388. [DOI] [PubMed] [Google Scholar]
- Dudley R (2004) Ethanol, fruit ripening, and the historical origins of human alcoholism in primate frugivory. Integr Comp Biol 44:315–323. [DOI] [PubMed] [Google Scholar]
- Dudley R (2014) The drunken monkey: why we drink and abuse alcohol. 1 ed., University of California Press. [Google Scholar]
- Dudley R (2020) The natural biology of dietary ethanol, and its implications for primate evolution, in Alcohol and Humans (HOCKINGS KJ, DUNBAR R eds), pp 9–23, Oxford University Press, Oxford, U.K. [Google Scholar]
- Duester G, Farrés J, Felder MR, Holmes RS, Höög JO, Parés X, Plapp BV, Yin SJ, Jörnvall H (1999) Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem Pharmacol 58:389–395. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC (2007) Genomic screen for substance dependence and body mass index in southwest California Indians. Genes, Brain and Behavior 6:184–191. [DOI] [PubMed] [Google Scholar]
- Ersoy A, Kelley J, Andrews P, Alpagut B (2008) Hominoid phalanges from the middle Miocene site of Paşalar, Turkey. Journal of Human Evolution 54:518–529. [DOI] [PubMed] [Google Scholar]
- Ferré P, Foufelle F (2007) SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Hormone Research in Paediatrics 68:72–82. [DOI] [PubMed] [Google Scholar]
- Gochman SR, Brown MB, Dominy NJ (2016) Alcohol discrimination and preferences in two species of nectar-feeding primate. R Soc Open Sci 3:160217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SM, Meijer RI, Barrett EJ (2014) Insulin regulates brain function, but how does it get there? Diabetes 63:3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman A, Säll T, Compagno C, Piskur J (2013) Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS One 8:e68734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbalch SG, Knudsen GM, Videbaek C, Pinborg LH, Schmidt JF, Holm S, Paulson OB (1999) No effect of insulin on glucose blood-brain barrier transport and cerebral metabolism in humans. Diabetes 48:1915–1921. [DOI] [PubMed] [Google Scholar]
- Hockings KJ, Bryson-Morrison N, Carvalho S, Fujisawa M, Humle T, McGrew WC, Nakamura M, Ohashi G, Yamanashi Y, Yamakoshi G, Matsuzawa T (2015) Tools to tipple: ethanol ingestion by wild chimpanzees using leaf-sponges. Royal Society Open Science 2:150150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N (2005) Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309:938–940. [DOI] [PubMed] [Google Scholar]
- Ingram LON, Buttke TM (1985) Effects of alcohols on micro-organisms, in Advances in Microbial Physiology, Vol. 25, Advances in Microbial Physiology (ROSE AH, TEMPEST DW eds), pp 253–300, Academic Press. [DOI] [PubMed] [Google Scholar]
- Johnson RJ, Andrews P (2010) Fructose, uricase, and the back-to-Africa hypothesis. Evol Anthropol 19 250–257. [Google Scholar]
- Johnson RJ, Andrews P (2015) The fat gene: A genetic mutation in prehistoric apes may underlie today’s pandemic of obesity and diabetes. Scientific American 313:64–69. [Google Scholar]
- Johnson RJ, Gaucher EA, Sautin YY, Henderson GN, Angerhofer AJ, Benner SA (2008) The planetary biology of ascorbate and uric acid and their relationship with the epidemic of obesity and cardiovascular disease. Med Hypotheses 71:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RJ, Sánchez-Lozada LG, Andrews P, Lanaspa MA (2017) Perspective: a historical and scientific perspective of sugar and its relation with obesity and diabetes. Adv Nutr 8:412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RJ, Stenvinkel P, Andrews P, Sánchez-Lozada LG, Nakagawa T, Gaucher E, Andres-Hernando A, Rodriguez-Iturbe B, Jimenez CR, Garcia G, Kang DH, Tolan DR, Lanaspa MA (2020) Fructose metabolism as a common evolutionary pathway of survival associated with climate change, food shortage and droughts. J Intern Med 287:252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RJ, Titte S, Cade JR, Rideout BA, Oliver WJ (2005) Uric acid, evolution and primitive cultures. Semin Nephrol 25:3–8. [DOI] [PubMed] [Google Scholar]
- Kelley J (2008) Identification of a single birth cohort in Kenyapithecus kizili and the nature of sympatry between K. kizili and Griphopithecus alpani at Pasalar. J Hum Evol 54:530–537. [DOI] [PubMed] [Google Scholar]
- Kelley J, Andrews P, Alpagut B (2008) A new hominoid species from the middle Miocene site of Paşalar, Turkey. J Hum Evol 54:455–479. [DOI] [PubMed] [Google Scholar]
- King T, Aiello LC, Andrews P (1999) Dental microwear of Griphopithecus alpani. J Hum Evol 36:3–31. [DOI] [PubMed] [Google Scholar]
- Kratzer JT, Lanaspa MA, Murphy MN, Cicerchi C, Graves CL, Tipton PA, Ortlund EA, Johnson RJ, Gaucher EA (2014) Evolutionary history and metabolic insights of ancient mammalian uricases. Proc Natl Acad Sci U S A 111:3763–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanaspa MA, Cicerchi C, Garcia G, Li N, Roncal-Jimenez CA, Rivard CJ, Hunter B, Andres-Hernando A, Ishimoto T, Sánchez-Lozada LG, Thomas J, Hodges RS, Mant CT, Johnson RJ (2012a) Counteracting roles of AMP deaminase and AMP kinase in the development of fatty liver. PLoS ONE 7:e48801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanaspa MA, Sánchez-Lozada LG, Choi YJ, Cicerchi C, Kanbay M, Roncal-Jimenez CA, Ishimoto T, Li N, Marek G, Duranay M, Schreiner G, Rodriguez-Iturbe B, Nakagawa T, Kang DH, Sautin YY, Johnson RJ (2012b) Uric acid induces hepatic steatosis by generation of mitochondrial oxidative stress: potential role in fructose-dependent and -independent fatty liver. J Biol Chem 287:40732–40744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanaspa MA, Sánchez-Lozada LG, Cicerchi C, Li N, Roncal-Jimenez CA, Ishimoto T, Le M, Garcia GE, Thomas JB, Rivard CJ, Andres-Hernando A, Hunter B, Schreiner G, Rodriguez-Iturbe B, Sautin YY, Johnson RJ (2012c) Uric acid stimulates fructokinase and accelerates fructose metabolism in the development of fatty liver. PLoS One 7:e47948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wanga j, Rosengberg D, Zhao Z, Lengyeld G, Nadele D (2018) Fermented beverage and food storage in 13,000 y-old stone mortars at Raqefet Cave, Israel: Investigating Natufian ritual feasting. J Archeol Science: Reports 21:783–793. [Google Scholar]
- Lustig RH (2013) Fructose: it’s “alcohol without the buzz”. Adv Nutr 4:226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenpaa PH, Raivio KO, Kekomaki MP (1968) Liver adenine nucleotides: fructose-induced depletion and its effect on protein synthesis. Science 161:1253–1254. [DOI] [PubMed] [Google Scholar]
- Marshall AJ, Wrangham R (2007) Evolutionary consequences of fallback foods. Int J Primatol 28:1219–1235. [Google Scholar]
- McCrossin ML, Benefit BR (1997) On the relationships and adaptations of Kenyapithecus, a large-bodied hominoid from the Middle Miocene of eastern Africa, in Function, Phylogeny and Fossils: Miocene Hominoid Evolution and Adaptations, Function, Phylogeny and Fossils: Miocene Hominoid Evolution and Adaptations, (BEGUN DR, WARD CV, ROSE MD eds), pp 241–268, Plenum Press, New York. [Google Scholar]
- McGovern P, Jalabadze M, Batiuk S, Callahan MP, Smith KE, Hall GR, Kvavadze E, Maghradze D, Rusishvili N, Bouby L, Failla O, Cola G, Mariani L, Boaretto E, Bacilieri R, This P, Wales N, Lordkipanidze D (2017) Early Neolithic wine of Georgia in the South Caucasus. Proc Natl Acad Sci U S A 114:E10309–E10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern PE, Zhang J, Tang J, Zhang Z, Hall GR, Moreau RA, Nunez A, Butrym ED, Richards MP, Wang CS, Cheng G, Zhao Z, Wang C (2004) Fermented beverages of pre- and proto-historic China. Proc Natl Acad Sci U S A 101:17593–17598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel JV (1962) Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? American journal of human genetics 14:353–362. [PMC free article] [PubMed] [Google Scholar]
- Nunn AV, Bell JD, Guy GW (2009) Lifestyle-induced metabolic inflexibility and accelerated ageing syndrome: insulin resistance, friend or foe? Nutr Metab (Lond) 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Satta Y, Takenaka O, Takahata N (2002) Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol Biol Evol 19:640–653. [DOI] [PubMed] [Google Scholar]
- Orowan E (1955) The origin of man. Nature 175:683–684. [DOI] [PubMed] [Google Scholar]
- Peana AT, Sánchez-Catalán MJ, Hipólito L, Rosas M, Porru S, Bennardini F, Romualdi P, Caputi FF, Candeletti S, Polache A, Granero L, Acquas E (2017) Mystic acetaldehyde: the never-ending story on alcoholism. Frontiers in Behavioral Neuroscience 11:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Morley A (2014) An evolutionary perspective on the Crabtree effect. Front Mol Biosci 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirola RC, Lieber CS (1972) The energy cost of the metabolism of drugs, including ethanol. Pharmacology 7:185–196. [DOI] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C (2006) How did Saccharomyces evolve to become a good brewer? Trends Genet 22:183–186. [DOI] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Müller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C (2011) Comparative functional genomics of the fission yeasts. Science 332:930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozpędowska E, Hellborg L, Ishchuk OP, Orhan F, Galafassi S, Merico A, Woolfit M, Compagno C, Piskur J (2011) Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts. Nat Commun 2:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE (2002) Segmental duplications and the evolution of the primate genome. Nat Rev Genet 3:65–72. [DOI] [PubMed] [Google Scholar]
- Sánchez F, Korine C, Pinshow B, Dudley R (2004) The possible roles of ethanol in the relationship between plants and frugivores: first experiments with egyptian fruit bats. Integr Comp Biol 44:290–294. [DOI] [PubMed] [Google Scholar]
- Sánchez-Lozada LG, Andres-Hernando A, Garcia-Arroyo FE, Cicerchi C, Li N, Kuwabara M, Roncal-Jimenez C, Johnson RJ, Lanaspa M (2019) Uric acid activates aldose reductase and the polyol pathway for endogenous fructose production and fat accumulation in the development of fatty liver. J Biol Chem in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaquist ER, Damberg GS, Tkac I, Gruetter R (2001) The effect of insulin on in vivo cerebral glucose concentrations and rates of glucose transport/metabolism in humans. Diabetes 50:2203–2209. [DOI] [PubMed] [Google Scholar]
- Skinner MF, Dupras TL, Moya-Sola S (1995) Periodicity of linear enamel hypoplasia among Miocene Dryopithecus from Spain. J Paleopathol:195–222. [Google Scholar]
- Softic S, Gupta MK, Wang GX, Fujisaka S, O’Neill BT, Rao TN, Willoughby J, Harbison C, Fitzgerald K, Ilkayeva O, Newgard CB, Cohen DE, Kahn CR (2018) Divergent effects of glucose and fructose on hepatic lipogenesis and insulin signaling. J Clin Invest 128:1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Softic S, Meyer JG, Wang GX, Gupta MK, Batista TM, Lauritzen H, Fujisaka S, Serra D, Herrero L, Willoughby J, Fitzgerald K, Ilkayeva O, Newgard CB, Gibson BW, Schilling B, Cohen DE, Kahn CR (2019) Dietary sugars alter hepatic fatty acid oxidation via transcriptional and post-translational modifications of mitochondrial proteins. Cell Metab 30:735–753 e734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengell M, Kubera B, Peters A (2021) Brain more resistant to energy restriction than body: a systematic review. Frontiers in Neuroscience 15:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavric B, Johnson WJ, Clayman S, Gadd RE, Chartrand A (1976) Effect of fructose administration on serum urate levels in the uricase inhibited rat. Experientia 32:373–374. [DOI] [PubMed] [Google Scholar]
- Steiper ME, Young NM (2006) Primate molecular divergence dates. Mol Phylogenet Evol 41:384–394. [DOI] [PubMed] [Google Scholar]
- Sussman RW, Tab Rasmussen D, Raven PH (2013) Rethinking primate origins again. Am J Primatol 75:95–106. [DOI] [PubMed] [Google Scholar]
- Tapia E, Cristobal M, Garcia-Arroyo FE, Soto V, Monroy-Sanchez F, Pacheco U, Lanaspa MA, Roncal-Jimenez CA, Cruz-Robles D, Ishimoto T, Madero M, Johnson RJ, Sánchez-Lozada LG (2013) Synergistic effect of uricase blockade plus physiological amounts of fructose-glucose on glomerular hypertension and oxidative stress in rats. Am J Physiol Renal Physiol 304:F727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke R (2019) Microsomal ethanol-oxidizing system: success over 50 years and an encouraging future. Alcoholism: Clinical and Experimental Research 43:386–400. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA (2005) Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet 37:630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffney BH (1984) Seed size, dispersal syndromes, and the rise of the angiosperms: evidence and hypothesis. Annals of the Missouri Botanical Garden 71:551–576. [Google Scholar]
- Van den Berghe G (1986) Fructose: metabolism and short-term effects on carbohydrate and purine metabolic pathways. Progress in biochemical pharmacology 21:1–32. [PubMed] [Google Scholar]
- Wang M, Chen WY, Zhang J, Gobejishvili L, Barve SS, McClain CJ, Joshi-Barve S (2020) Elevated fructose and uric acid through aldose reductase contribute to experimental and human alcoholic liver disease. Hepatology 72:1617–1637. [DOI] [PubMed] [Google Scholar]
- Wiens F, Zitzmann A, Lachance MA, Yegles M, Pragst F, Wurst FM, von Holst D, Guan SL, Spanagel R (2008) Chronic intake of fermented floral nectar by wild treeshrews. Proc Natl Acad Sci U S A 105:10426–10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Moriwaki Y, Takahashi S (2005) Effect of ethanol on metabolism of purine bases (hypoxanthine, xanthine, and uric acid). Clin Chim Acta 356:35–57. [DOI] [PubMed] [Google Scholar]
- You M, Arteel GE (2019) Effect of ethanol on lipid metabolism. J Hepatol 70:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]


