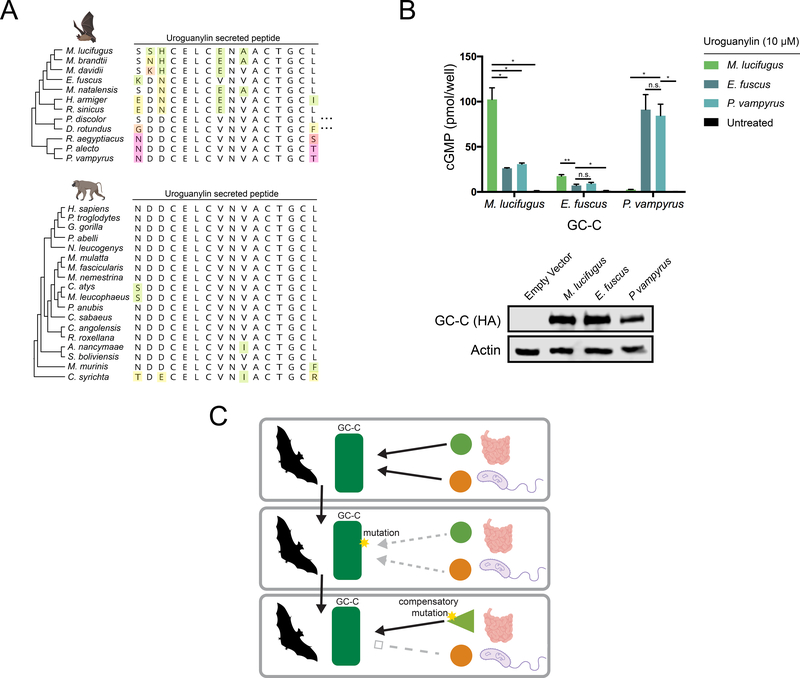
Figure 4: Coevolution of GC-C and uroguanylin in bats.
(A) Amino acid alignment of the mature uroguanylin peptide based on genome sequences of the indicated bat (top) or primate (bottom) species. Residues with mutations relative to the consensus within each clade are uniquely colored. Elipses represent stop codon loss mutations in the indicated bat species. (B) (Top) cGMP levels in HEK293T cells expressing P. vampyrus or M. lucifugus GC-C after treatment with 10 μM uroguanylin from the indicated species. * indicates a p-value < 0.05 and ** indicates a p-value<0.01 using one-way ANOVA followed by Dunnett’s T3 multiple comparison test. (Bottom) immunoblot of HA-tagged GC-C from the indicated cell lines. (C) Model for pathogen driven evolution of uroguanylin in bats. Mutations in GC-C that result in a loss of affinity for both STa and uroguanylin may provide a net fitness benefit, allowing time for compensatory mutations to arise.