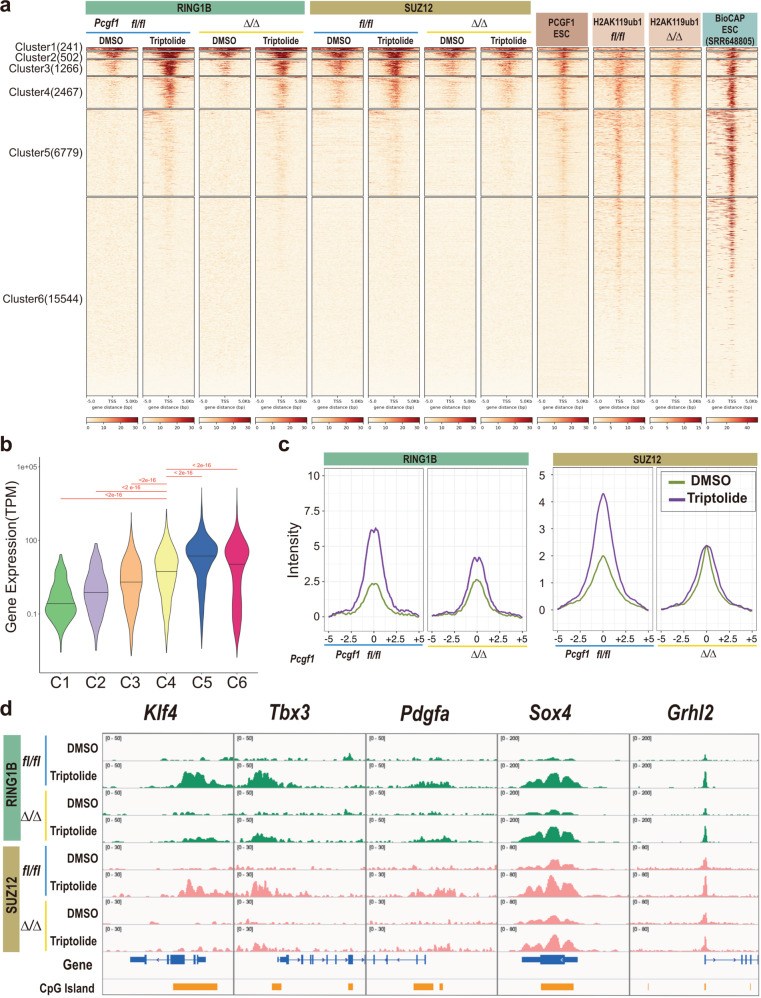
Fig. 4. Inhibition of RNA polymerase II facilitates PRC2 accumulation in a group of genes in a PCGF1-dependent manner.
a Heatmap views for RING1B and SUZ12 distribution at the TSS (5 kb upstream to 5 kb downstream) of each gene in WT (fl/fl) or Pcgf1-KO (∆/∆) ESCs, with or without (DMSO) Triptolide treatment. In parallel, the distribution of PCGF1 in untreated control ESCs, and H2AK119ub1 enrichment in control (fl/fl) or Pcgf1-KO (∆/∆) ESCs without Triptolide treatment, are shown as a reference. BioCAP data (SRR648805 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1064680], deposited in GEO) showing the distribution of unmethylated CpGs are also shown. Genes were sorted into six groups by k-means clustering (n = 6) based on the distribution of RING1B, SUZ12, and PCGF1 in WT (fl/fl) ESCs. The number of genes in each cluster is shown within parentheses. b Violin plot showing differential gene expression in each cluster in untreated ESCs. Significant differences were calculated by Mann–Whitney U test (one-sided). The p-values are adjusted by BH methods. c Average binding of RING1B and SUZ12 in Cluster 4 genes in WT or Pcgf1-KO ESCs and EBs. Metaplot views of RING1B and SUZ12 binding at the TSS (5 kb upstream to 5 kb downstream) of Cluster 4 genes, in WTl (fl/fl) or Pcgf1-KO (∆/∆) ESCs, with or without triptolide treatment. Data for triptolide-treated, or untreated, ESCs are shown by purple and green lines, respectively. d Distribution of RING1B and SUZ12 at selected Group 1 genes (Klf4, Tbx3, Pdgfa) and Group 3 genes (Sox4, Grhl2) in control ESCs with or without (DMSO) Triptolide treatment. Genomic snapshots of RING1B and SUZ12 distributions in WT (fl/fl) or Pcgf1-KO (∆/∆) ESCs with or without (DMSO) triptolide treatment are shown.