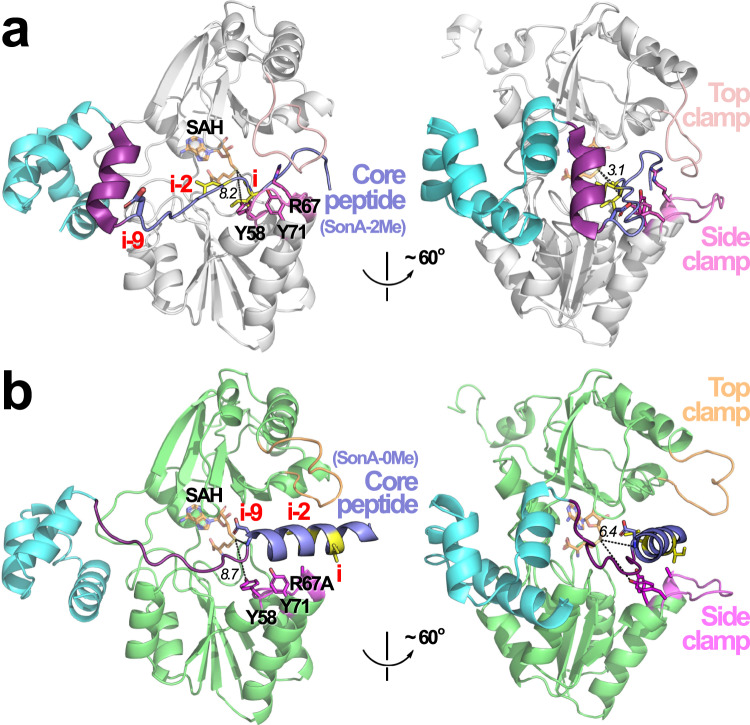
Fig. 7. Global differences in structures of SonM—SonA-2Me—SAH and SonM-R67A—SonA-0Me—SAH complexes.
a One SonM—SonA-2Me—SAH heterodimer is shown, where SonM is represented in white cartoon, the BBD of SonA-2Me is shown in cyan cartoon, and the core peptide is shown in slate. α-N-Methylated residues “i” and “i–2” are depicted as yellow sticks. Helix 5 of the BBD is highlighted purple. The two structures in this panel are rotated ~60 degrees to view the structure from two different perspectives. b One SonM-R67A—SonA-0Me—SAH heterodimer is shown, where SonM is represented in light green cartoon, and all other portions of the structure are colored and depicted as in a. Note that residues “i” and “i−2” are not methylated in this structure as SonM-R67A is an inactive mutant. Key distances are depicted as black dashed lines and their lengths noted in italics in Ångstroms. In comparison to a, the SonM-R67A—SonA-0Me—SAH complex reveals a pre-reacted state, where the top lock and clamps are open and the core peptide is more than 3 Å farther away from the cofactor. More significantly, the core peptide is α-helical and positions deeper into the active site, resulting in the BBD helix 5 being unwound into a coil.