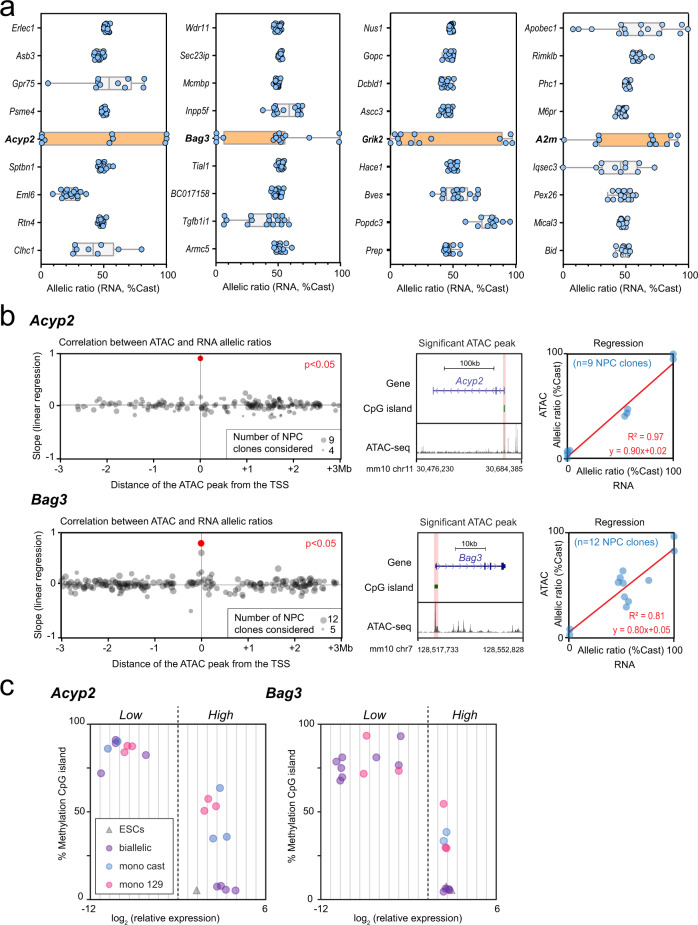
Fig. 7. Allelic imbalance is regulated locally.
a Comparison of the distribution of allelic expression ratios for Acyp2, Bag3, Grik2 and A2m, with four upstream and four downstream neighbouring genes, measured by allele-specific RNA-seq in 16 NPC clones. Boxplot elements: centre line represents the median, box bounds represent the 25th and 75th percentiles, whiskers represent the minimum and maximum. The orange colour highlights the boxplot plot of the RME gene of interest. b Identification of the genomic region where allele-specific accessibility correlates best with expression, by linear regression between the ATAC-seq allelic accessibility ratio and RNA-seq allelic expression ratio, for ATAC-seq peaks (containing at least one SNP) within a region of ±3 megabases around the TSS. P value adjusted with Bonferroni correction, number of ATAC peaks: 177/207 Acyp2/Bag3 (left panels). UCSC genome browser snapshot showing a representative ATAC-seq track along the gene, with highlighting of the ATAC-seq peak, located at the TSS, that correlates significantly with the RNA-seq (middle panels). Linear regression showing the correlation between the ATAC-seq allelic accessibility ratio and the RNA-seq allelic expression ratio at the TSS (right panels). ATAC-seq data and RNA-seq datasets are from 13 NPC clones. Acyp2 F(1,7) = 227, p = 1.3 × 10−6; Bag3 F(1,9) = 42, p = 7.3 × 10−5. c DNA methylation levels over the CpG island located at the TSS of Acyp2 and Bag3, measured by Sequenom bisulfite analysis in 21 representative NPC clones and the two ES cell lines, characterised by high or low expression levels and mono- or biallelic expression.