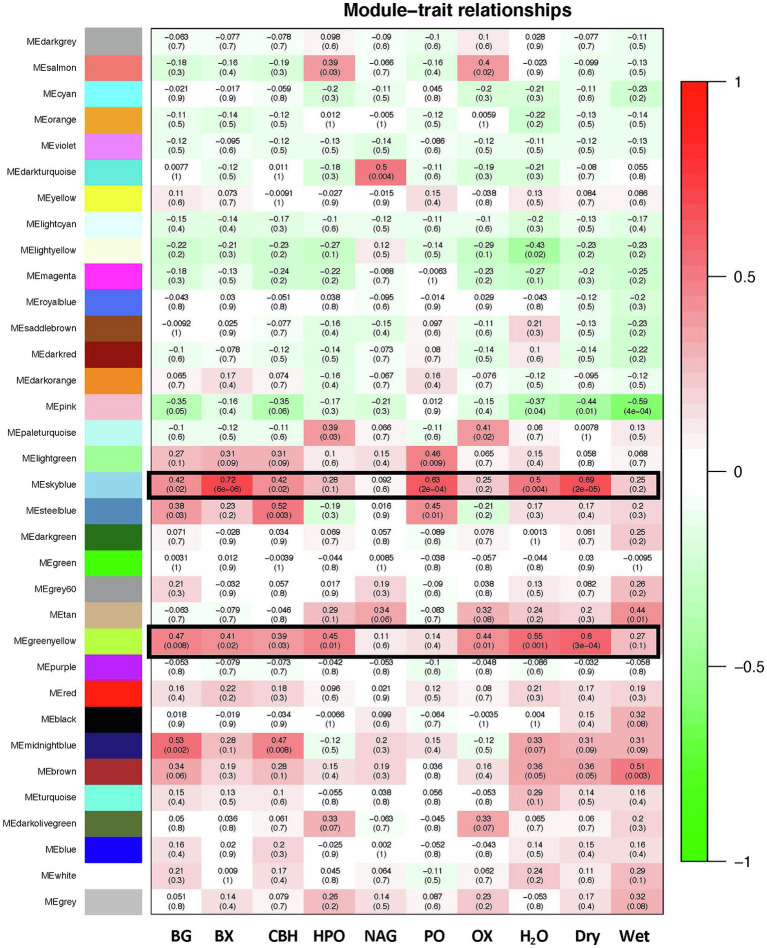
Figure 5.
Co-weighted network analyses – module-trait associations between KEGG annotated transcript abundances and enzyme concentrations and physical soil parameters. Each row corresponds to a module eigen gene and each column represents a trait. The top number indicates the correlation (|cor|) between the eigen gene value and the individual trait and the bottom number indicates the p-value. The colors from green to red indicate positive or negative correlation, respectively. Enzyme codes – BG, β-glucosidase; BX, β-xylosidase; CBH, cellobiohydrolase; HPO, peroxidase; NAG, β-N-acetyl-glucosaminidase; PO, phenol oxidase; dry, dry weight of samples; and wet, wet weight of samples. Trait data are taken from Pold et al. (2017).