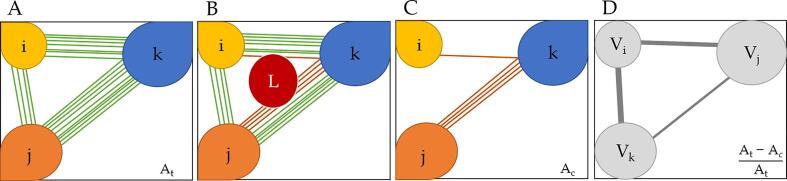
Fig. 1.
Extraction and modelling of a disconnectome. A. Simplified representation of the atlas-based tractogram connectivity of three brain regions i, j and k and their respective streamlines. At is the associated adjacency matrix of the atlas connectome, where each element At(i,j) represents the number of streamlines connecting the pair of regions (i,j). B. Overlapping of the atlas tractogram with the lesion mask. Streamlines passing through the lesion L are highlighted in red. C. Affected streamlines are isolated. Ac is the adjacency matrix of the affected streamlines, where each element Ac(I,j) represents the number of affected streamlines connecting the pair of regions (i,j) D. The brain graph representation where brain areas i, j and k are represented respectively by nodes Vi Vj and Vk and edges are weighted by the relative number of affected streamlines. The adjacency matrix of the remaining connectivity is defined as the relative difference between At and Ac. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)