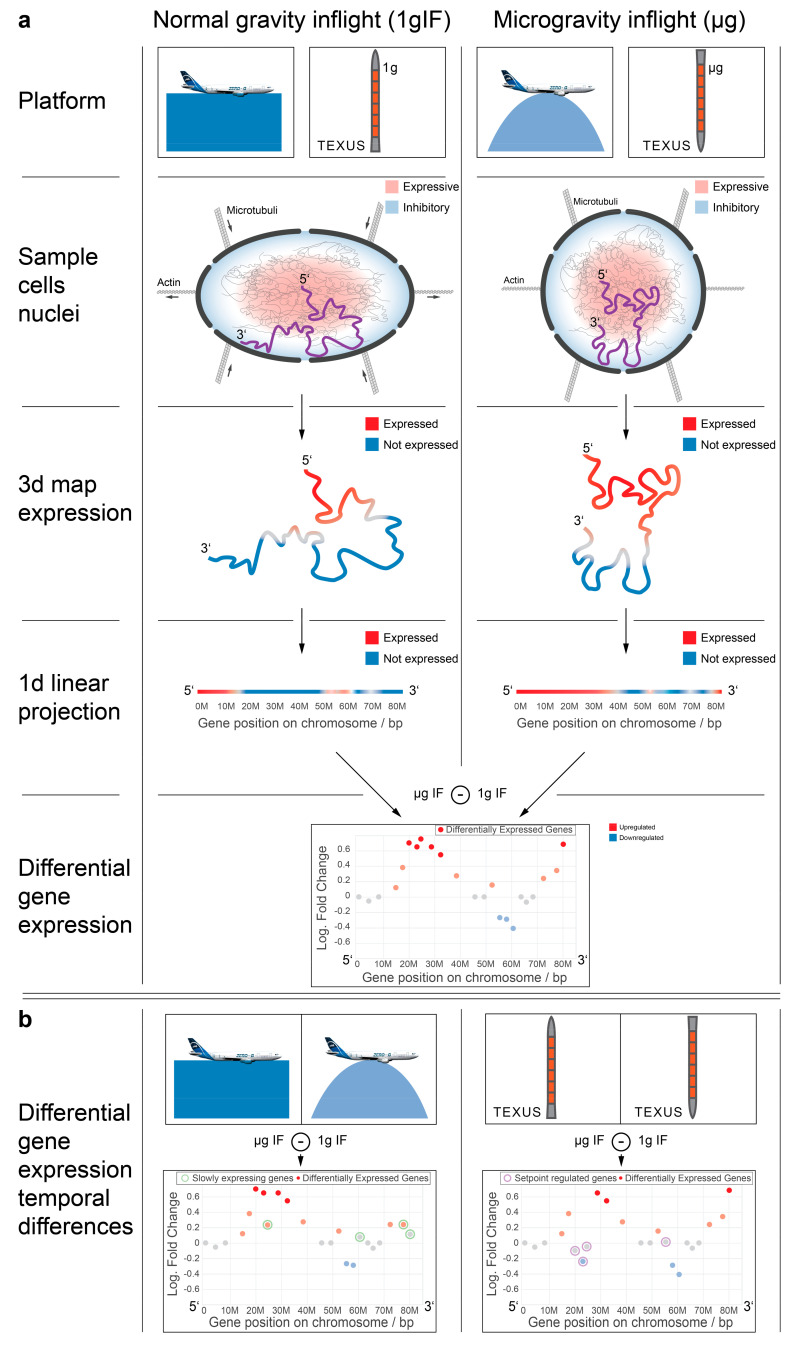
Figure 9.
Summary of the proposed hypothesis. (a) The effects of altered gravity on cellular gene expression are proposed to have a mechanical nature. Cells on different platforms are exposed to both normal gravity and microgravity. During the normal gravity, the forces acting by gravity slightly compress the nucleus in the vertical axis, therefore shortening it in the vertical axis and prolonging it in the horizontal axis. In microgravity, these forces are no longer present, and the nucleus takes a more roundish shape. Resulting from different nuclear shapes, the chromatin has to take different orientations, depending on the shape. An exemplary chromosome, highlighted in purple, takes different conformations, depending on the platform flight phase. Different areas of the nucleus are known to exhibit different expressivities, the outer areas are rather repressive, and the inner areas are rather expressive. This leads to different expression strengths for different genes, depending on where the gene is located on the chromosome and therefore in the nucleus. When projecting this three-dimensional (3D) to one-dimensional conformation, the distinct expression/repression pattern also becomes visible in one dimension. Because of the conformation change upon altered gravity, there are regional differences in expressivity, if regions are no longer in same areas. When subtracting both patterns from each other during transcriptomics studies, this leads to regional patterns of fold changes distributions of differentially expressed genes. (b) Differential gene expression datasets from different platforms are not equal on the gene level, since slowly expressing genes only move to their distinct regional level after a longer period of time (not given during a parabolic flight; green circles), and next to mechanogenetic effects, further gene regulatory effects will influence the expression patterns, for example setpoint regulation bringing back upregulated genes to target levels after a while (purple circles). This explains why differential expression sets from similar conditions on different platforms differ a lot, even if they have the same underlying effect. This disturbance effect on the level of single genes can be compensated for by the GRCR analysis described here.