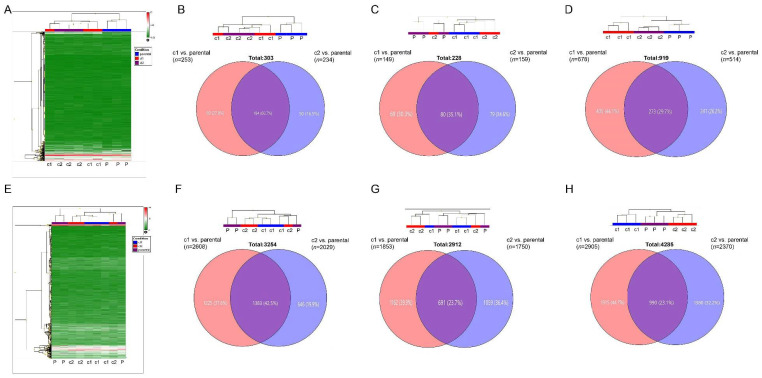
Figure 2.
Microarray analysis of gene and miRNA expression in resistant and parental control cell lines: (A) Representative unsupervised cluster analysis of miRNA expression (786-O cells). (B–D) (Top) Dendrogram of unsupervised cluster analysis showing relationship between samples at level of miRNA expression; (Bottom) Venn diagram depicting differentially expressed miRNAs in resistant clones relative to parental control cells. (B) 786-O cells, (C) A498 cells, (D) Caki-1 cells. (E) Representative unsupervised cluster analysis of gene expression (786-O cells). (F–H) (Top) Dendrogram of unsupervised cluster analysis showing relationship between samples at level of gene expression; (Bottom) Venn diagram depicting differentially expressed genes in resistant clones relative to parental control cells. (F) 786-O cells, (G) A498 cells, (H) Caki-1 cells. The original heatmap analyses are shown in Figure S4. Differentially expressed miRNAs and genes that are common between the resistant clones are listed in (Tables S1–S6).