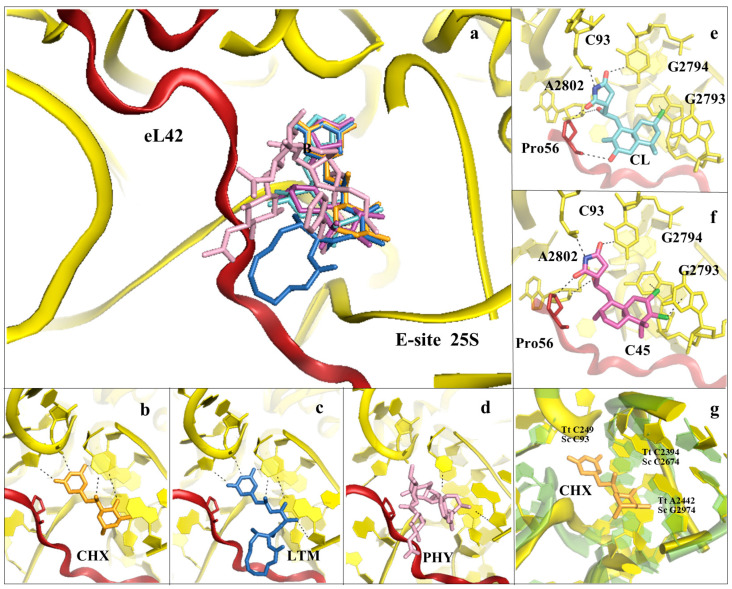
Figure 5.
Inhibitors binding to the E-site of the LSU impair the translocation process. (a) Overview of the inhibitors targeting specifically the eukaryotic E-site. Cycloheximide (CHX) (PDB: 4U43) in orange, lactimidomycin (LTM) in blue (PDB: 4U4R), phyllantoside (PHY) in pink (PDB: 4U4Z), chlorolissoclimide (CL) in light cyan (PDB: 5TBW) and the synthetic analogue C45 in magenta (PDB: 6HHQ); (b) zoom-in and details of interaction for CHX within the E-site binding pocket; (c) zoom-in and details of interaction for LTM within the E-site binding pocket; (d) zoom-in and details of interaction for PHY within the E-site binding pocket. PHY, in contrast to CHX and LTM, is reported to interact directly with the eukaryotic-specific ribosomal protein eL42. eL42 is coloured in red for clarity; (e) zoom-in of CL interaction network within the E-site, with details of the hydrogen bonding and the peculiar halogen–π interaction that lissoclimides establish with rRNA residues. Additionally, CL interacts with the backbone of eL42; (f) same view for C45, with highlighted the second halogen–π interaction that the drug establishes with the rRNA. This additional bond might compensate for the lack of the hydrogen bond with eL42, as it occurs with CL. In both images, the type of bonding established between the drug and the ribosome is marked; (g) structural superimposition of the S. cerevisiae LSU (coloured in yellow) with the LSU of T. thermophilus (coloured in light green) (PDB: 4V5D) (rmsd 4.5 Å). Numbers of residues for the two organisms are shown (Sc: S. cerevisiae, Tt: T. thermophilus).