Figure 4.
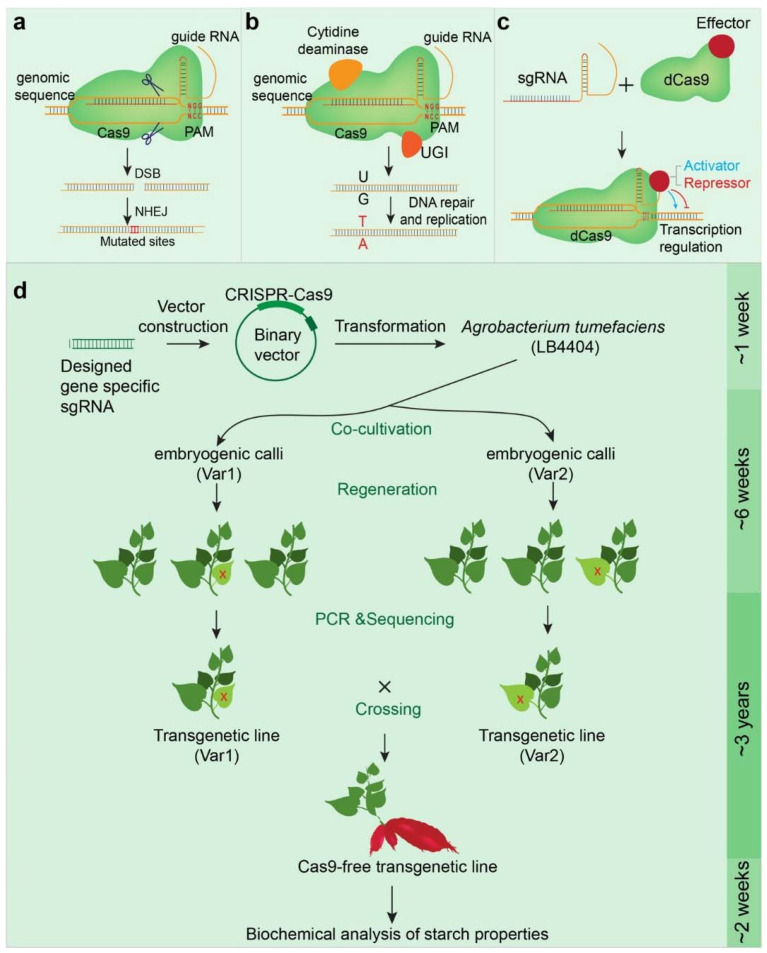
The multifunctional CRISPR/Cas9 system and a schematic description of sweet potato genome-editing and selection for T-DNA-free progeny. (a) The basic CRISPR/Cas9 system. The designed guide RNA, based on a specific target DNA sequence, binds to the target site, and directs the Cas9 protein to the genomic sequence complementary to sgRNA, adjacent to a protospacer adjacent motif (PAM). PAM comprises three nucleotides “NGG”, where n represents any nucleotide. Cas9 acts as the endonuclease that cuts the DNA sequence specifically recognized by the sgRNA. Upon cleavage, the genomic DNA forms double-strand breaks (DSBs). The DSBs are then repaired by non-homologous end joining (NHEJ), a conserved mechanism that repairs the DSB in the genome. (b) Base editing technology. Cytidine deaminase is fused to an inactivated Cas9 protein to generate a Cas9 nickase (nCas9) that acts as a cytosine base editor (CBE). CBE generates C·G to T·A base substitutions. UGI is the uracil DNA glycosylase inhibitor. The important function of UGI is to prevent mutagenesis by eliminating uracil from DNA molecules by cleaving the N-glycosidic bond and initiating the base-excision repair pathway. To generate A·T to G·C substitutions, the cytidine deaminase is replaced by an adenine deaminase to create an adenine base editor (ABE). (c) The CRISPR interference (CRISPRi). The inactivated dCas9 (endonuclease deficient Cas9), which lacks endonuclease activity but retains guide RNA binding activity, is fused with transcriptional effectors (activators or repressors). Upon binding to the promoter of a target gene, guided by a designed guide RNA, the effector activates (by an activator fusion) or represses (by a repressor fusion) the target gene expression without changing the DNA sequence. (d) A working flow illustrating genome editing and selection of T-DNA-free progeny of tuber crop plants, such as sweet potato [16], that are normally bred through tissue propagation rather than seed selection. In short, a binary vector (pCambia 1300) containing CRISPR/Cas and sgRNA is used to transform sweet potato mediated by Agrobacterium tumefaciens (LB4404). Due to the self-incompatibility of sweet potato, two varieties (Var1 and Var2) were transformed to generate calli. The regenerated seedlings were selected by PCR and DNA sequencing to identify transgenic lines (represented by dark green leaves) that are mutated at the target sites (red X). The two variety transgenic lines were then crossed to generate segregating F1 hybrids that contain the mutation, but with or without the T-DNA (Cas9 and guide RNA). Only the lines with the mutation but without T-DNA (-Cas9) are selected and moved forward for subsequent characterization. CRISPR/Cas has shown significant applicability in genome editing-based crop breeding and will have a profound impact on the future of agriculture [33,120]. CRISPR technology accelerates the process of plant breeding and is continuously evolving. The base editors and primer editors, recently developed on the CRIPSR/Cas platform, enable precise mutations of single nucleotides, or deletions and insertions of DNA fragments in a genomic position [121,122]. Gene editing is also advantageous when multiple genes must be mutated. For instance, the inactivation of both SBEIIa and SBEIIb in wheat reduced amylose by more than 70%. However, no changes in amylose content is detected when only SBEIIa or SBEIIb is mutated [123]. Simultaneously mutating multiple genes can be achieved by CRISPR/Cas [124].