Figure 1.
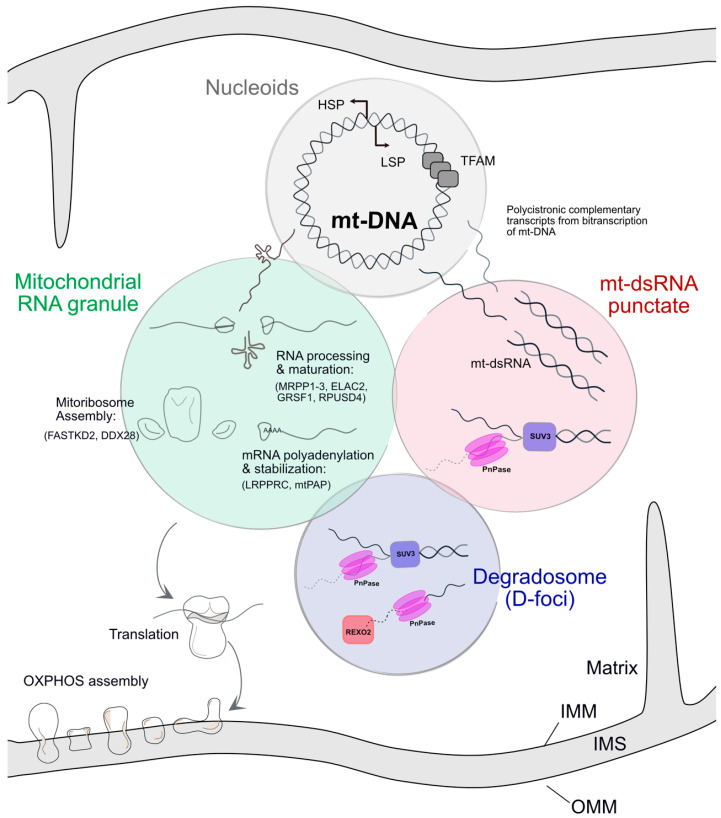
Nucleoids containing a single mt-DNA molecule, condensed by TFAM, transcribe polycistronic mRNAs. A single-stranded mRNA transcript is thought to undergo endonucleolytic processing and maturation in the Mitochondrial RNA granule (MRG). It is also the site of mitoribosome assembly. Matured mRNA transcripts transit out of the MRGs to be translated into subunits of the ETC. Bidirectional transcription of the mt-DNA from its Heavy strand promoter (HSP) and Light strand promoter (LSP) can lead to the complementary transcripts forming duplex mt-dsRNA molecules which have been observed in foci similar to MRGs. mt-dsRNA is mainly regulated by the degradosome proteins, SUV3-PNPase. The Degradosome (D-foci) is the site of mt-RNA degradation by SUV3, PNPase and REXO2. While colocalization between Nucleoids, MRGs and D-foci has been demonstrated, the association of mt-dsRNA granules to these structures has yet to be further investigated. Either they are separate entities or part of the MRGs, which would imply that the different structures are tightly connected. IMM: Inner mitochondrial membrane, IMS: Intermembrane space, OMM: Outer mitochondrial membrane.