Figure 5.
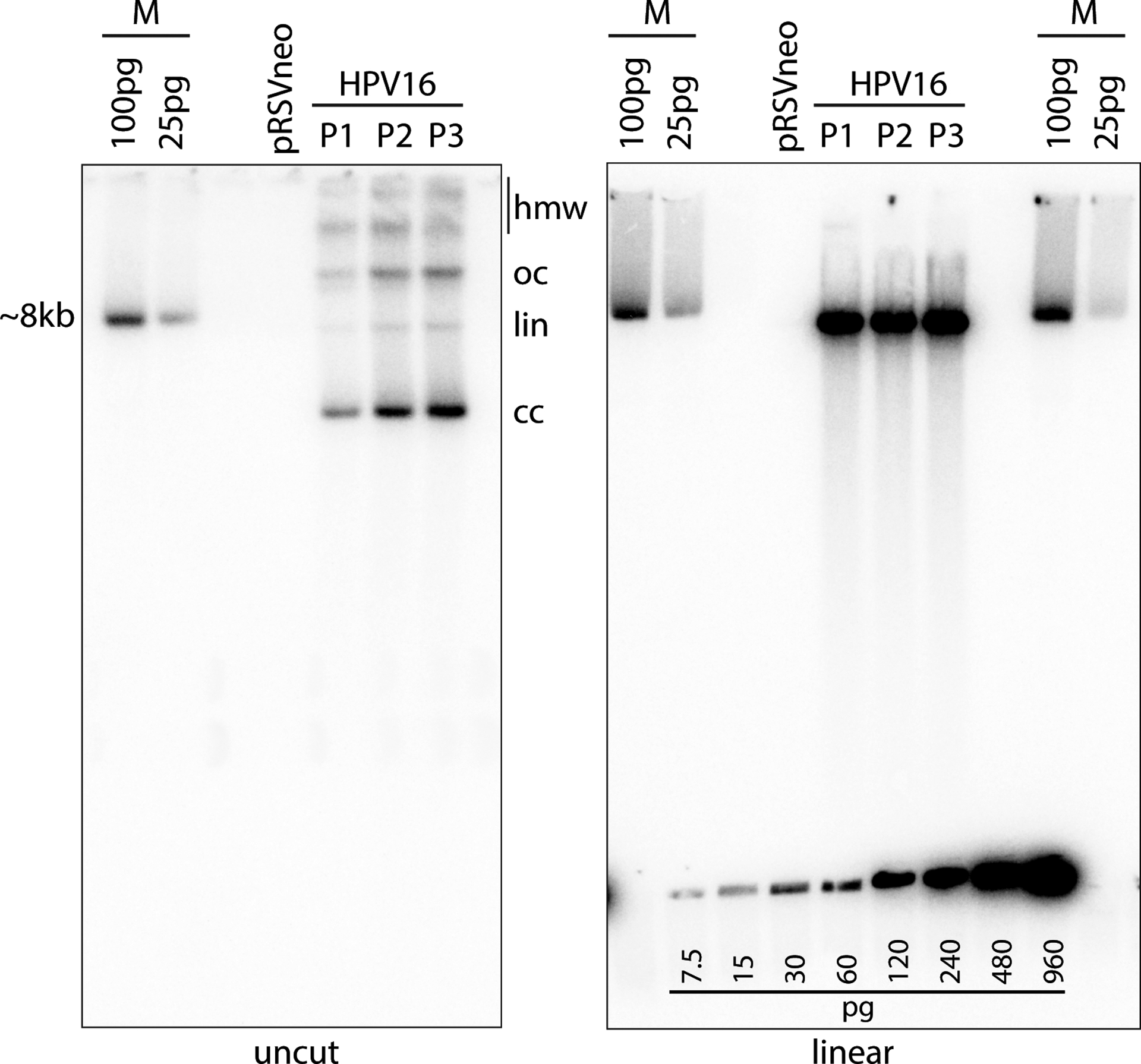
Southern blot analysis. In this example, DNA was extracted from cells passed from the colonies shown in Figure 2 using the Qiagen DNeasy Blood and Tissue Kit (Support Protocol 4). Three micrograms of total cellular DNA was digested with Hindlll (cleaves cellular DNA but does not cut HPV16 viral genome) or BamHl (cuts the viral genome once) and analyzed by Southern blot analysis as detailed in Basic Protocol 3. The left gel shows the HPV DNA isomers detected by the radioactive probe (see Supporting Protocol 3): supercoiled, covalently closed (cc), linear (lin) DNA, open circular/nicked (oc) DNA, and high molecular weight (hmw) forms that may include concatemers. Linearized viral DNA digested with BamHl runs as a single ~8kb band as shown in the gel on the right. The cloned linear HPV16 genome, cleaved from the bacterial vector, is used as a size marker (M). A DNA standard was also created using the HPV16 size marker and was loaded along the bottom of the gel on the right. This DNA standard can be used to estimate the viral DNA copy number. The calculated loaded DNA standard amount should be adjusted to account for the cloned bacterial plasmid and compared to the viral DNA signal in the cell DNA lane. The amount of DNA in a single human cell is ~6pg. Therefore, 3μg cell DNA represents 5 × 105 cells. Each pg of viral DNA (7900bp dsDNA) contains 1.155 ×105 molecules. Thus, the number of viral DNA copies per cell can be easily calculated.
