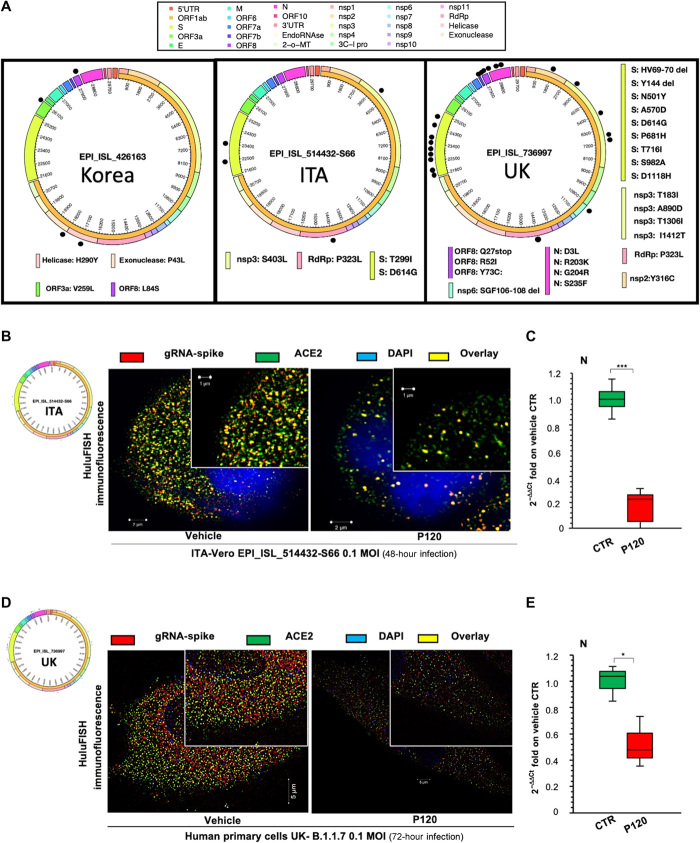
Fig. 3. polyP120 exerts antiviral action against SARS-CoV-2 variants belonging to 20A and 20I/501Y.V1 (B.1.1.7) clades.
(A) Circos plot showing missense variations among 19A clade (EPI_ISL_426163), 20A clade (EPI_ISL_514432-S66), and 20I/501Y.V1 (B.1.1.7) clade (EPI_ISL_736997) SARS-CoV-2 variants used in this work. Each black dot represents a missense mutation in the viral genomes. (B to D) HuluFISH analyses. (B and D) HuluFISH analysis with a pan–SARS-CoV-2 probe against the S gene (red) coupled to immunofluorescence (IF) staining with an antibody against the ACE2 protein (green). (B) Vero E6 cells were infected with Italian SARS-CoV-2 particles (MOI, 0.1) for 24 hours and then were treated with 37.5 μM polyP120 for additional 24 hours. (D) Primary human epithelial cells from nasal brushing were pretreated with 37.5 μM polyP120 for 1 hour and then infected with Italian SARS-CoV-2 particles (MOI, 0.1) for 72 hours. SARS-CoV-2–infected cells treated with vehicle (CTR) were used as negative controls in (B) and (D). The SIM image was acquired with Elyra 7 and processed with Zeiss ZEN software (blue edition). Magnification, ×63. (C and E) Quantification of mRNA abundance relative to that in the CTR (2−ΔΔCt) of genomic viral RNAs (N gene) from RT-PCR analysis of total RNA extracted from Vero E6 cells infected with Italian SARS-CoV-2 (EPI_ISL_514432-S66) (C) or primary human epithelial cells from nasal brushing infected with B.1.1.7 SARS-CoV-2 (UK) (EPI_ISL_736997) (E). SARS-CoV-2–infected cells treated with vehicle (CTR) were used as controls. Data are means ± SD. *P < 0.05 and ***P < 0.001 by unpaired two-tailed Student’s t test, adjusted with the Bonferroni method; n = 3 independent experiments per group. DAPI, 4′,6-diamidino-2-phenylindole.