Figure 3.
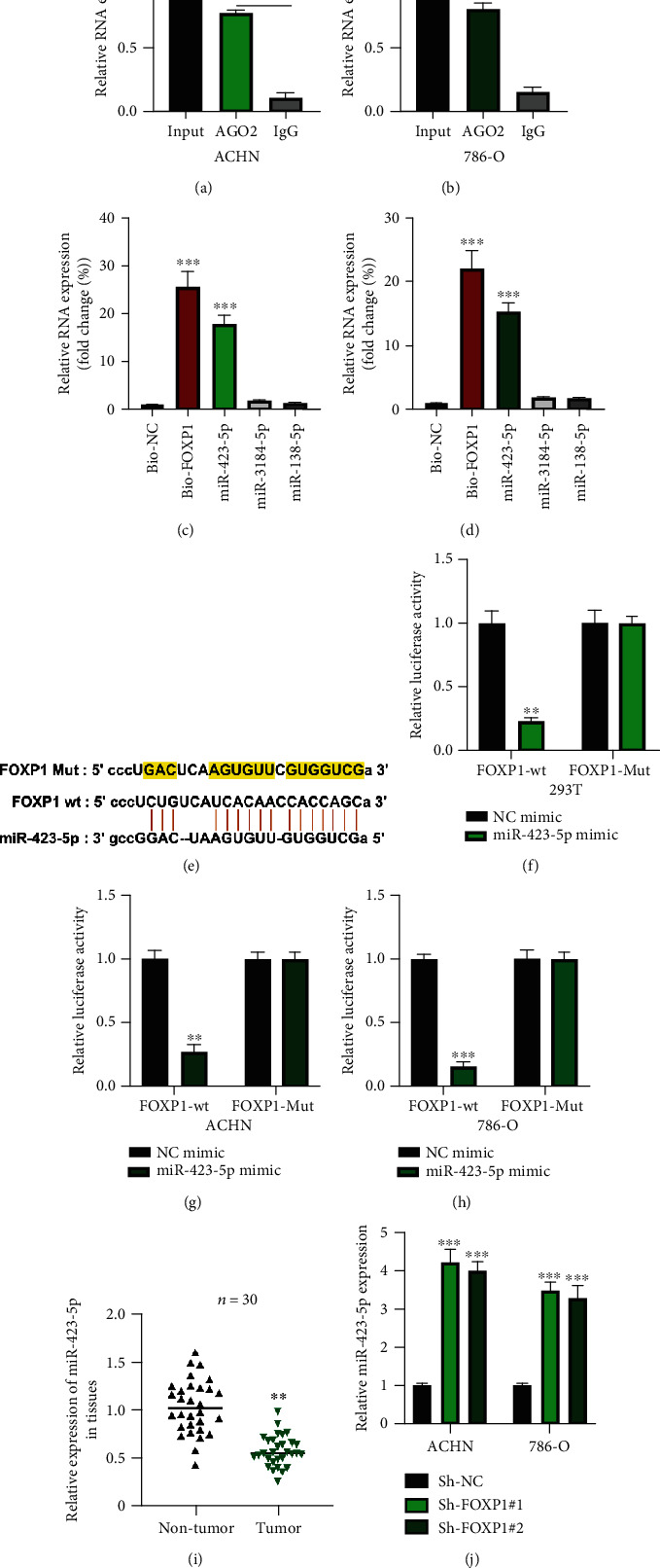
circFOXP1 sponges to miR-423-5p. (a, b) AGO2-RIP was utilized in ACHN (a) and 786-O (b) cells; relative circFOXP1 was measured by qRT-PCR. (c, d) Bioinformatics prediction of circFOXP1 targets was performed using the ENCORI dataset (http://starbase.sysu.edu.cn/) with CLIP Data: medium stringency (≥2), and Degradome Data: low stringency ≥1). Results were tested using biotinylated RNA pull-down; relative RNA expression was measured by qRT-PCR in ACHN (c) and 786-O cells (d). (e) Predicted circFOXP1 wild type (wt) or mutant type (Mut) bind sequences with miR-423-5p. (f–h) Relative luciferase activities in reporter vector-treated 293T (f), ACHN (g), and 786-O (h) cells were measured as indicated. (i) Expression level of miR-423-5p in thirty pairs of RCC tissues was determined by qRT-PCR assay. (j) Expression level of miR-423-5p in circFOXP1 downregulated ACHN, and 786-O cells were measured by qRT-PCR. All assays were conducted at least three times, ∗∗P < 0.01 and ∗∗∗P < 0.001.
