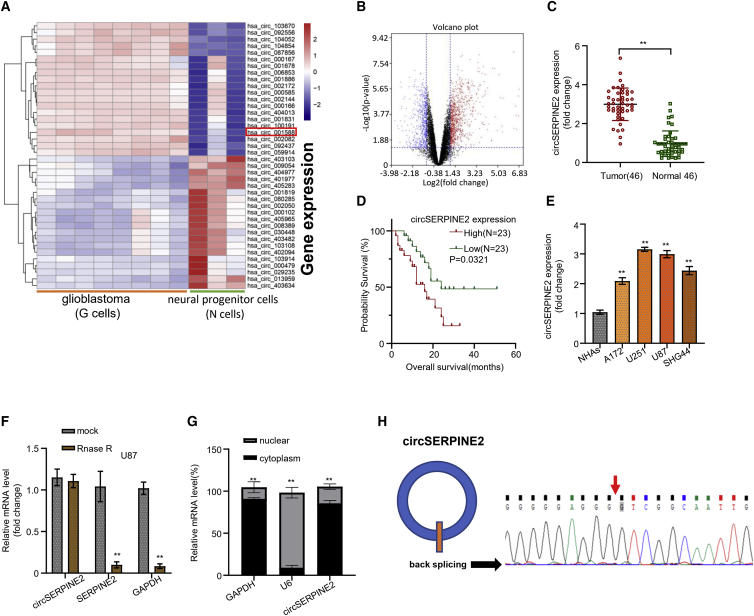
Figure 1.
circSERPINE2 is upregulated in glioblastoma tissue and cells
Heatmap produced from hierarchical clustering of 1,361 differentially expressed circRNAs in 8 GM cells and 3 neural progenitor (N) cells; the most upregulated and downregulated circRNAs are represented in red and purple respectively (A). Volcano plot of differentially expressed circRNAs showed that a total of 822 and 539 circRNAs were significantly upregulated and downregulated, respectively; black is the no differential expression, blue is significantly downregulated, and red is upregulated (B). Quantitative real-time PCR detects that circSERPINE2 expression is markedly upregulated in GM cancer tissues compared to normal brain tissues (C). Survival curve of GM patients in the high- and low circSERPINE2 expression group showed that high circSERPINE2 expression level was significantly associated with the poor prognosis of GM patients (D). Quantitative real-time PCR analysis of circSERPINE2 expression level in normal human astrocytes (NHAs) and GM cell lines (A172, U251, U87, and SHG44); circSERPINE2 was markedly upregulated in the GM cell lines with the highest expression level in the U251 and U87 cell lines (E). RNase R treatment to determine the circular nature circSERPINE2 in GM cell line; Rnase R had no significant effect on the expression level of circSERPINE2 mRNA when compared to the blank control mock (F). Detection of the subcellular localization of circSERPINE2; quantitative real-time PCR analysis showed that circSERPINE2 was significantly expressed in the cytoplasmic fraction of the GM cell line compared to the nuclear fraction (result is comparable to that of the nuclear-enriched U6 and cytoplasmic-enriched GAPDH) (G). Sequencing of junction sites of circSERPINE2 (H). ∗∗p < 0.01.