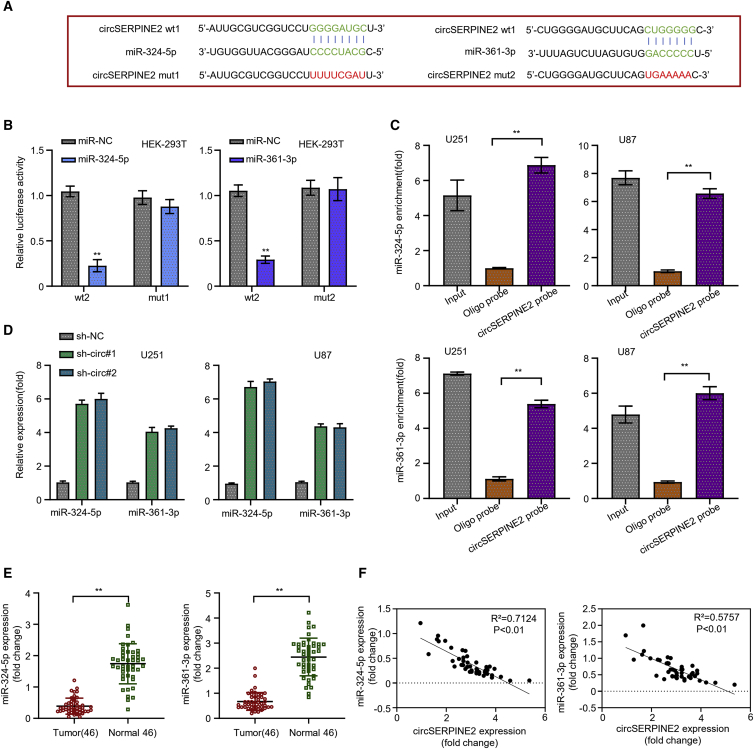
Figure 3.
circSERPINE2 sponges miR-361-3p and miR-324-5p and accelerates their degradation
The circular RNA interactome predicted the complementary sequence of circSERPINE2 as miR-361-3p and miR-324-5p (A). Luciferase reporter assay experiment in HEK293T cells co-transfected with miR-361-3p and miR-324-5p (mut and WT) and miR-NC; the luciferase activity of the HEK293T cells transfected with circSERPINE2 WT sequence was significantly reduced but no significant inhibiting effect was observed in those transfected with the circSERPINE2 mut sequence (B). Biotinylated RNA pull-down assay revealed that more miR-361-3p and miR-324-5p mRNA were enriched in the circSERPINE2 oligonucleotide probe in the GM cell lines compared to the control oligo probe (C). Quantitative real-time PCR analysis showed that circSERPINE2 knockdown significantly upregulated miR-361-3p and miR-324-5p expression level in GM cell lines (D). Differential expression analysis of miR-361-3p and miR-324-5p mRNA in 46 GM cancer tissues compared to the normal brain tissues; miR-361-3p and miR-324-5p was significantly downregulated in the GM tissues (E). Correlation analysis of miR-361-3p or miR-324-5p expression level and circSERPINE2 expression level; their expression was negatively correlated (F). ∗∗p < 0.01.