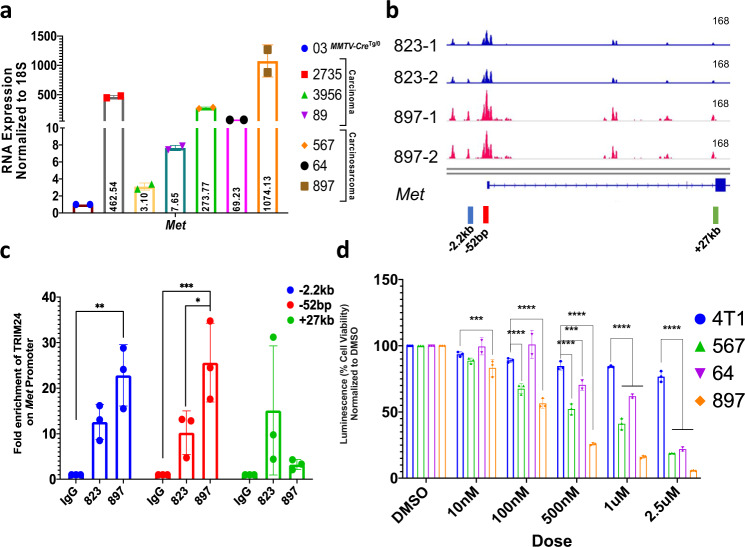
Fig. 5. TRIM24 directly activates c-Met to support metaplastic carcinosarcoma cell viability.
a qRT-PCR validation of cMET RNA expression in TRIM24 overexpressing tumors compared to MMTV-CreTg/0 mammary gland (03) (n = 2 technical replicates for each tumor samples). b Snapshot of Integrated Genome Viewer (IGV) showing tracks and peaks of ATAC-Seq-determined chromatin accessibility of MMTV-Cre spontaneous tumor cell line 823 (blue) and TRIM24 overexpressing primary metaplastic carcinosarcoma cell line 897 (red). The scales of the tracks are adjusted to 168. The color bar below the tracks shows positions of primers used to assess TRIM24 interactions with cMET chromatin. c Chromatin immunoprecipitation (ChIP) PCR showing enrichment of TRIM24 on Met promoter to validate ATAC-Seq on 823 and 897 cell lines. The color of each bars indicates primers corresponding to arrows shown in B (n = 3 biological replicates). (*p = 0.0209, **p = 0.0014, ***p = 0.0004). d Cell viability assay on 4T1 (mouse TNBC cell line- non-metaplastic) and TRIM24 overexpressing primary cell lines (567, 64, and 897) upon treatment with Crizotinib, an FDA-approved ATP-competitive selective small molecule inhibitor of c-Met and ALK receptor. Percent cell viability is measured using a luminescence reader; values are normalized to DMSO, a vehicle control (n = 3 biological replicates). Data represented as mean with SD in a, and mean ± SEM in c, d. p-values are calculated in c based on two-way ANOVA for multiple comparisons using Tukey method and in d using two-way ANOVA for multiple comparisons using Holm–Sidak method. (***p < 0.001, ****p < 0.0001).