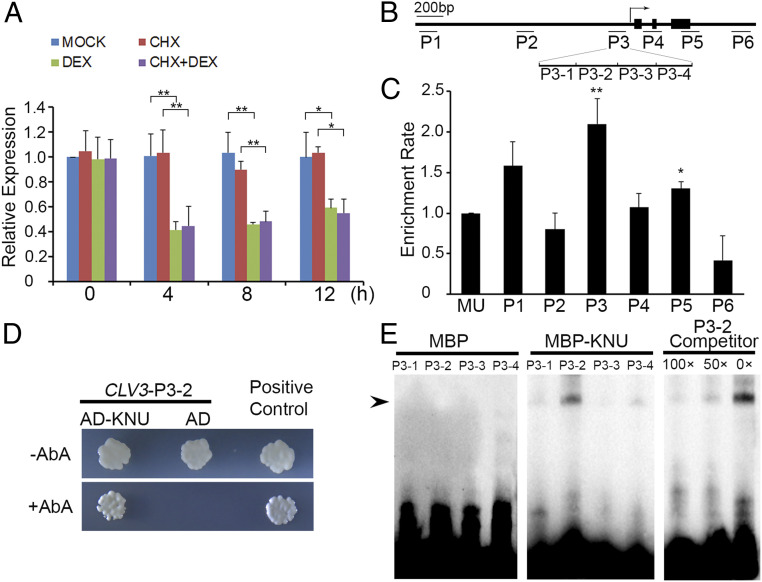
Fig. 2.
KNU directly represses CLV3. (A) CLV3 expression levels in ap1 cal 35S:KNU-GR-myc after single DEX treatment, CHX treatment, and DEX + CHX treatment. CLV3 transcript levels were quantified by qPCR. Tip41-like gene (At4g34270) served as the internal control. The error bars represent SD of three biological replicates. The asterisks indicate significant differences between samples treated with different chemicals (*P < 0.05 and **P < 0.01, Student’s t test). (B) Schematic diagram of CLV3 locus and primer sets P1 to P6 used for ChIP assays. (C) ChIP assay using ap1 cal 35S:KNU-GR-myc inflorescences. Nuclear proteins were immunoprecipitated with anti–c-Myc agarose beads, and the enriched DNA was used for qPCR assays. The y-axis shows relative enrichment compared with no antibody (negative control). Mu-like transposon (MU) served as a negative control locus, and the values of MU were calibrated to 1. The error bars represent SD of three biological replicates. The asterisks indicate significant differences between MU and different primer sets on CLV3 (*P < 0.05 and **P < 0.01, Student’s t test). (D) Yeast one-hybrid assays show that KNU interacts with the P3 region of CLV3. AbA, Aureobasidin A. (E) EMSAs confirm that KNU binds to the P3-2 fragment. The black arrow indicates the DNA–protein complex. Nonlabeled oligonucleotides were used as competitors. MBP was used as a negative control.