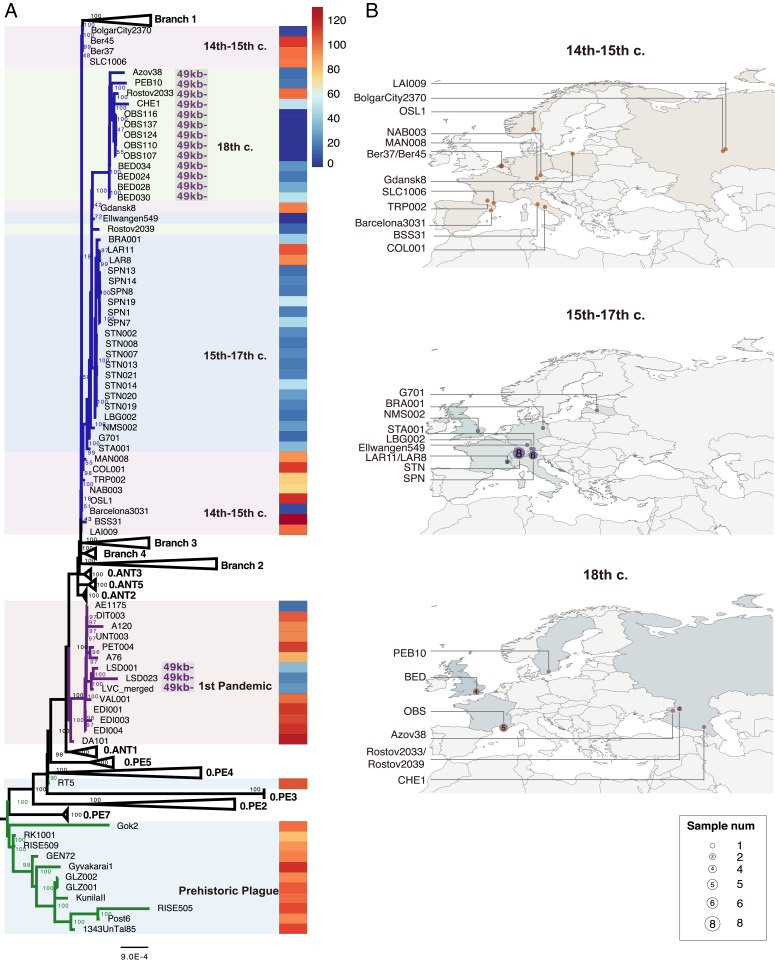
Fig. 1.
Phylogeny and archaeological site locations of ancient genomes. (A) A maximum likelihood phylogeny was obtained with 574 genomes of Y. pestis (including 75 ancient genomes) involved based on 12,608 SNPs. The numbers at each node indicate the bootstrap values of 1,000 replicates. Branches highlighted in blue correspond to the second pandemic, which is subdivided in three groups: the 14th to 15th century group, which also includes the Black Death and the Pestis secunda (1,357 to 1,366) strains; the 15th to 17th century group; and the 18th century group (which also includes the BED genomes for homogeneity). Branches in purple correspond to the first Pandemic, and branches in green correspond to the prehistoric plague. The ratio between the depth of pla and that of the entire pPCP1 plasmid for all ancient genomes is shown in the rightmost heatmap, with a color scale ranging from 0 (dark blue) to 130+ (dark red). (B) Geographic distribution of the three waves during the second pandemic.