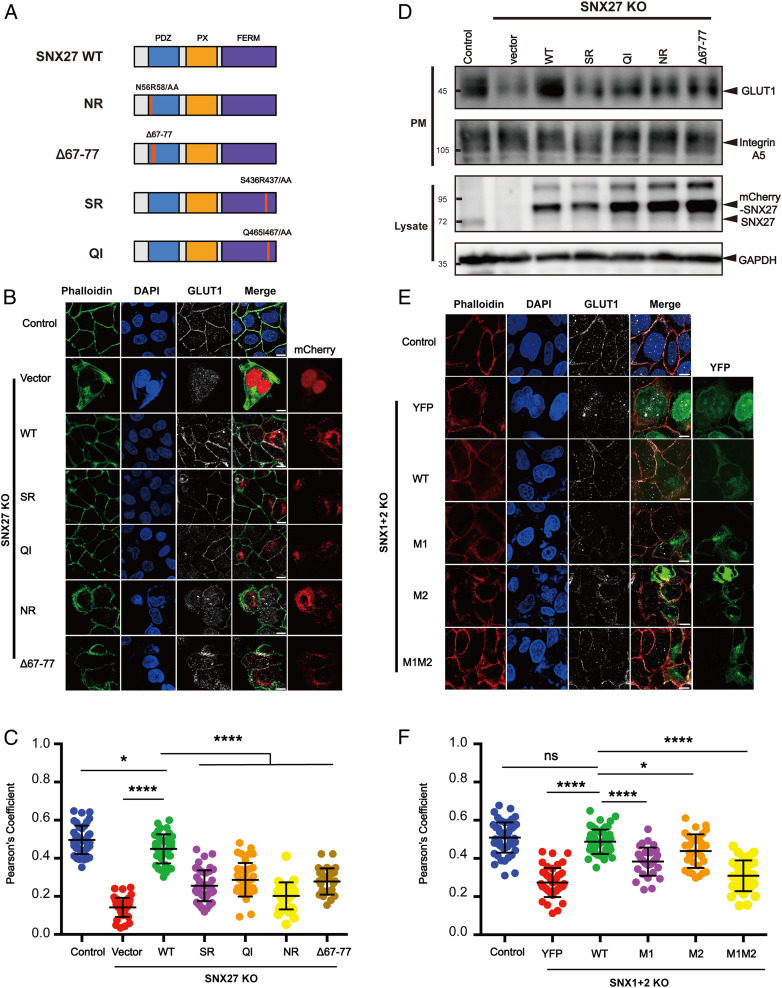
Fig. 5.
The interaction between SNX27 and SNX1 is critical for endosomal recycling of GLUT1. (A) A schematic diagram of SNX27 constructs used in this study. (B and C) Surface distribution of GLUT1 in HeLa cell lines. The SNX27 KO cells were transfected with mCherry, mCherry-SNX27 WT, mCherry-SNX27 SR, mCherry-SNX27 QI, mCherry-SNX27 NR, and mCherry-SNX27 Δ67 to Δ77 (Left), while control cells were left untreated. Cells were then incubated with antibody against GLUT1 (gray) and stained with phalloidin (green). (C) Colocalization analysis between GLUT1 and phalloidin in B. Each dot represents Pearson’s correlation coefficients from one cell. Data are presented as mean ± SD, and P values were calculated using one-way ANOVA and Tukey’s multiple comparisons test. *P < 0.05, 0.001 < ****P < 0.0001; figure is representative of n = 3 independent experiments with similar results. (D) Surface biotinylation level of GLUT1 and integrin A5 in HeLa cell strains. SNX27 KO cells were transfected with indicated plasmids in A (Top), while control cells were left untreated. Cells were then digested, washed, and subjected to biotin label reagent for 30 min at RT. Streptavidin agarose resin was utilized to capture biotinylated proteins. Samples were washed and used for immunoblotting (plasma membrane [PM]); mCherry-SNX27, endogenous SNX27, and GAPDH were detected in lysate. (E and F) Surface distribution of GLUT1 in HeLa cell lines. The SNX1+2 KO cells were transfected with YFP, YFP-SNX1 WT, YFP-SNX1 M1, YFP-SNX1 M2, and YFP-SNX1 M1M2 (Left), while control cells were left untreated. Cells were then incubated with antibody against GLUT1 (gray) and stained with phalloidin (red). (E) Colocalization analysis between GLUT1 and phalloidin in D. Each dot represents Pearson’s correlation coefficients from one cell. Data were presented as mean ± SD and P values were calculated using one-way ANOVA and Tukey’s multiple comparisons test. *P < 0.05, 0.001 < ****P < 0.0001, ns, not significant; figure is representative of n = 3 independent experiments with similar results.