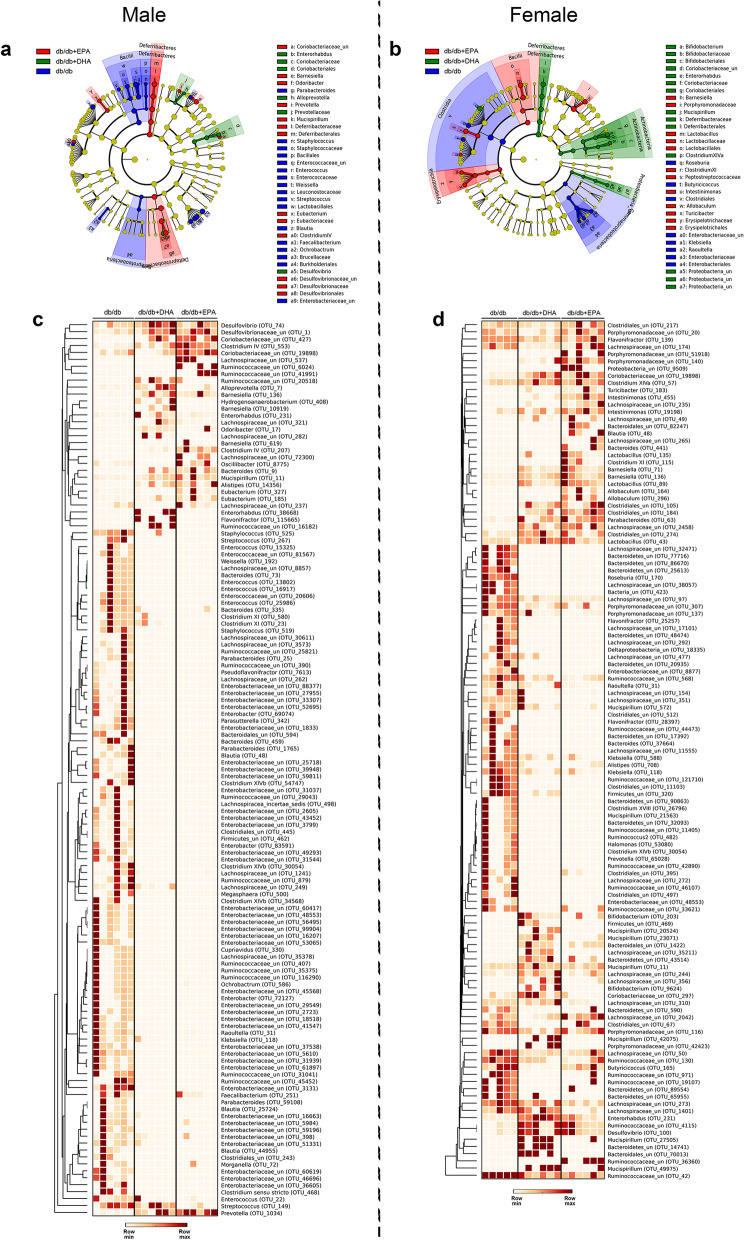
Fig. 2.
DHA and EPA differentially alter gut microbial phylotypes in db/db mice. Fecal microbiota composition in mice from different groups were analyzed using 16S rDNA sequencing (n = 6 male and n = 5-6 female per group). a, b Cladogram obtained from LEfSe analysis (LDA > 2), showing the most differentially abundant taxa enriched in microbiota from mice in db/db (blue), db/db + DHA (green), and db/db + EPA (red) groups. c, d Heatmap showing OTUs with significantly different relevant abundances between three treatment groups (as determined by Kruskall-Wallis test followed by Mann–Whitney test). Each row represents an OTU labeled by the lowest taxonomic description and OTU ID, normalized to the row maximum