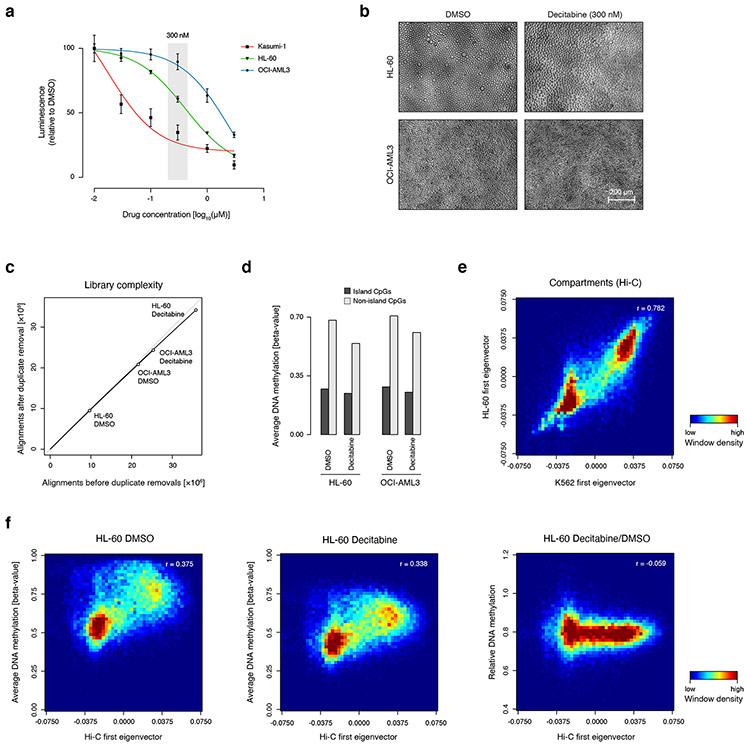
Extended Data Fig. 5 ∣. Characterization of decitabine treatment of cancer cell lines.
A) Plot shows dose response curve for decitabine treatment of three cell lines Kasumi, HL-60, and OCI-AML3. Viability was measured using cell titer glo and is reported as luminescence relative to control DMSO treated cells (n=3 independently treated replicates, error bars represent standard deviation).
B) Images show HL60 and OCI-AML3 cells treated with 300 nM decitabine and a DMSO vehicle control. Morphology of decitabine treated cells similar to control, repeated three times. Scale bar is indicated and applies to all images.
C) Plot shows unique reads as a function of aligned reads in XRBS libraries from DMSO- and decitabine-treated HL-60 and OCI-AML3 cells.
D) Barplot shows average DNA methylation values across island (dark grey) and non-island (light grey) CpGs in DMSO- and decitabine-treated HL-60 and OCI-AML3 cells. For example, average methylation of non-island CpGs in HL-60 cells is reduced from 68.1% to 54.3% by decitabine treatment (20.2% reduction, n=1 library per treatment).
E) Heatmap shows correlation between Hi-C-derived first eigenvectors from K562 and HL-60 cell lines in 100kb-windows, indicating high agreement in compartment structure between both cell lines.
F) Heatmaps show correlation between average DNA methylation values and Hi-C-derived eigenvector in 100kb-windows for DMSO- (left) and decitabine-treated HL-60 cells (center). Heatmap on the right shows relative DNA methylation values of decitabine- and DMSO-treated cells. Despite compartment B showing lower methylation compared to compartment A at baseline, induced DNA hypomethylation with decitabine treatment affects compartment A and B equally.