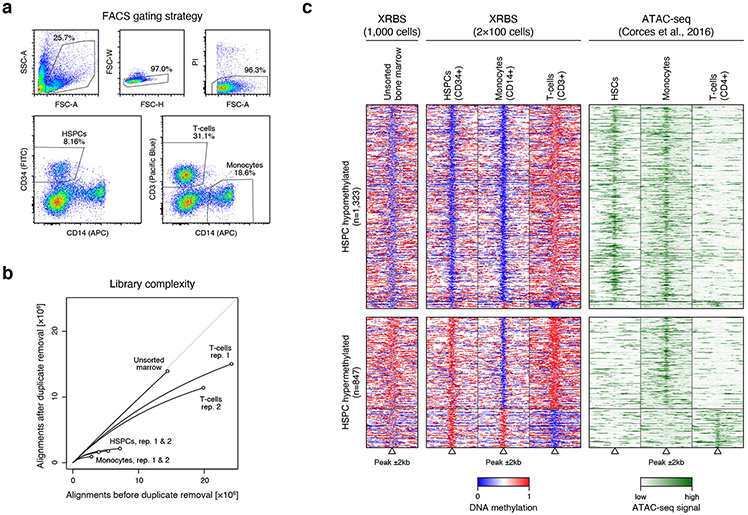
Extended Data Fig. 8 ∣. XRBS profiling of limited human bone marrow cell types.
A) Plots show the gating strategy for fluorescence assisted cell sorting (FACS) of human bone marrow of CD34+ HSPCs, CD3+ T cells, and CD14+ monocytes. Singlets (FSC-W vs. -H) and viable cells (PI vs. FSC-A) were sorted based on cell surface marker signal.
B) Plot shows unique reads as a function of aligned reads in libraries from unsorted human bone marrow, HSPCs, monocytes, and T cells. Libraries were generated from 100 sorted cells. 1000 cells were used for the unsorted bone marrow library.
C) Heatmap depicts 4kb regions (rows, n=2,170 regions) centered over elements defined in the ENCODE SCREEN database. Only differentially methylated elements between monocytes and T cells are shown. Elements were stratified by their methylation status in HSPCs (hypomethylated: top; hypermethylated: bottom). Methylation levels of the unsorted bone marrow are shown for comparison (left). ATAC-seq signal for sorted hematopoietic stem cells (HSCs), monocyte, and CD4+ T cells (obtained from 36).