Extended Data Fig. 9 ∣. Evaluation of single cell XRBS profiles.
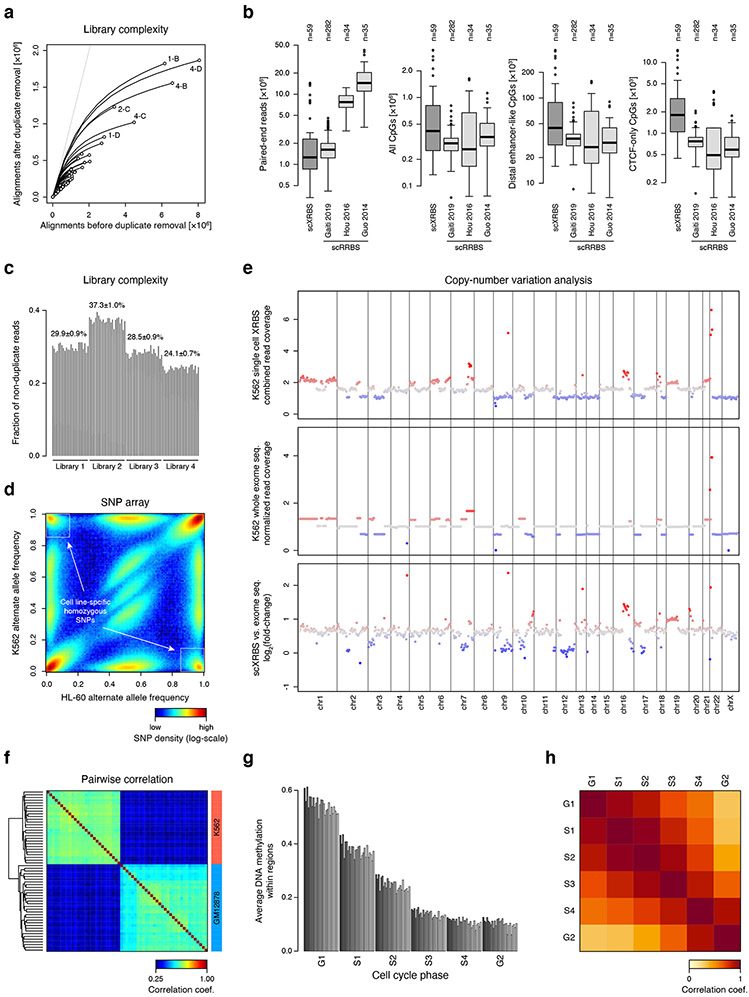
A) Plot shows unique reads as a function of aligned reads in single cell XRBS profiles (n=96 cells). With greater sequencing depth the fraction of unique reads decreases, as the chance of sampling a non-unique read (i.e. PCR duplicate) increases.
B) Boxplots compare DNA methylation profiles from human scXRBS (n=59 cells) and three published scRRBS datasets generated from human cells: Chronic lymphocytic leukemia (n=282 cells) 49, hepatocellular carcinoma and HepG2 cells (n=34 cells) 45, and oocytes, sperm and pronuclei (n=35 cells) 48. Single cells from Hou et al. were generated using the scTrio-seq protocol that in part resembles scRRBS. Only CpGs within 75 bases of an MspI cut site were considered for scRRBS libraries to adjust for differences in read lengths. Libraries from Gaiti et al. were sequenced at 2x51 bases. Left plot shows the number of paired-end reads sequenced for each cell. Other plots show the number of CpGs covered (≥1-fold) across all CpGs in the genome, CpGs within distal enhancer-like regions, and CpGs within ‘CTCF-only’ regions (SCREEN database). Both strands of a CpG dinucleotide are assessed individually. Although sequenced at the lowest depth, scXRBS libraries on average capture the most CpGs, particularly in CpG-sparse regions. Boxplots were generated in R using default settings: Bounds of box and thick horizontal line represent 25th, 75th, and 50th percentile of observations, whiskers represent minimum and maximum observations, and outliers are indicated as dots.
C) Barplot shows the fraction of unique reads (i.e. reads not representing PCR duplicates) per single cell library. Within the same PCR reaction, the duplicate rate was very similar, irrespective of the total number of aligned reads per single cell. Each bar plot represents a single cell XRBS library. Twenty four barcoded cells were in each of 4 independent libraries.
D) Heatmap compares alternate allele frequencies from SNP array data for K562 and HL-60 cell lines. Cell line-specific homozygous alleles are indicated by white boxes boxes and were used for single cell SNP analysis in Fig. 5d.
E) Plots show copy number variation calls from combined single cell XRBS profiles (top) and whole exome sequencing data (middle) for K562 cells. A number of chromosomes show differences in copy number between XRBS and whole exome sequencing (bottom). However, these differences likely represent true copy number variations between K562 cells used for these experiments.
F) Heatmap shows pairwise correlation coefficients of single cell methylation profiles. Dendrogram shows unsupervised clustering. Single cell XRBS profiles cluster by cell type.
G) Barplot shows K562 single cell average DNA methylation values within various early and late replicating regions. Each bar represents an individual K562 single cell library. There are 32 single cell libraries plotted for each cell cycle phase.
H) Heatmap shows pairwise correlation of average DNA methylation values within various early and late replicating regions. Late replicating regions (G2 phase) cluster separately. These results suggest that one source of single cell DNA methylation heterogeneity is decreased fidelity of maintenance DNA methylation in late replicating domains.