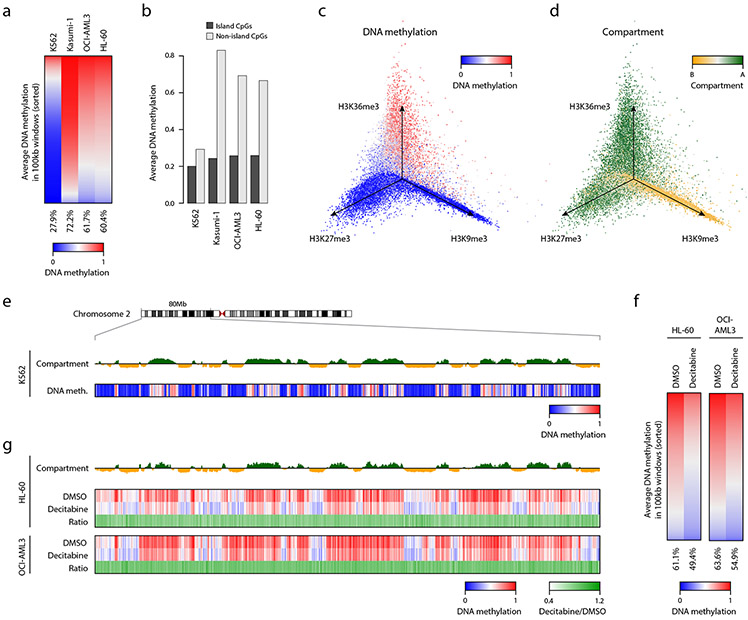
Fig. 3 ∣. XRBS reveals variable hypomethylation in cell lines and in response to demethylating agent.
A) Heatmap shows genome-wide DNA methylation in 100kb windows across four cell lines. Windows are sorted by decreasing DNA methylation for each cell line. Average methylation for each cell line is indicated below.
B) Bar plot shows average DNA methylation levels for CpG islands and non-CpG island regions across cell lines. Individual samples are shown.
C) Isometric projection plot positions all 100kb genomic windows (dots) according to their normalized signal for three histone modifications in K562 cells. Windows are colored by average DNA methylation of non-island CpGs. Windows marked by repressive marks (H3K27me3 and H3K9me3) are generally unmethylated, while only windows marked by H3K36me3 are methylated, consistent with very low global DNA methylation in K562 cells (see also Extended Data Fig. 4F).
D) Isometric projection plot as in panel C but with windows colored by the Hi-C experiment-derived first eigenvector 21. The H3K36me3 and H3K27me3 histone marks overlap with compartment A, while the H3K9me3 mark overlaps with compartment B in K562 cells.
E) Genome plot shows average DNA methylation and the Hi-C-derived first eigenvector for K562 cells for a 80Mb genomic segment of chromosome 2. Megabase-scale hypomethylated blocks primarily overlap compartment B (orange), whereas shorter hypomethylated regions and hypermethylated regions overlap compartment A (green).
F) Heatmap shows genome-wide DNA methylation in 100kb windows across DMSO and decitabine treated HL-60 and OCI-AML3 cells. Windows are sorted according to DNA methylation for each treatment. Average methylation for each cell line is indicated below.
G) Genome plot for the same region as in panel E shows the Hi-C-derived first eigenvector illustrating compartments A (green) and B (orange) for HL-60 cells 26, and average DNA methylation for HL-60 or OCI-AML3 treated with decitabine or control. Ratio shows methylation difference between decitabine and control. Decitabine reduces methylation in both compartments to a similar extent.