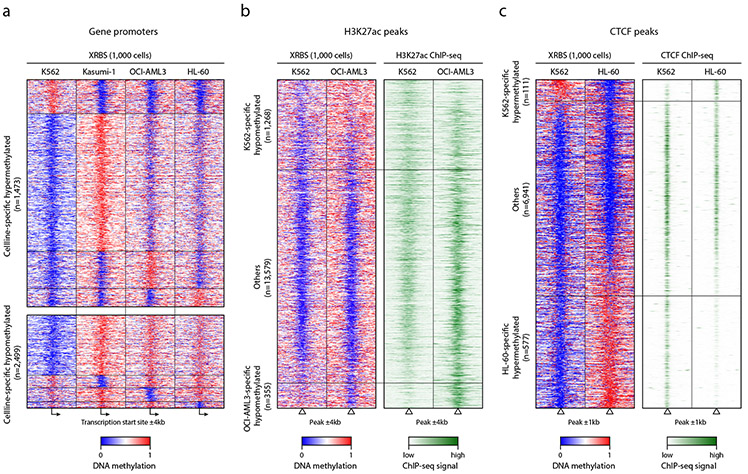
Fig. 4 ∣. XRBS predicts regulatory states of functional elements.
A) Heatmap depicts 8kb genomic regions (rows, n=3,972 promoters) centered at transcription start sites and divided into 100 equally sized bins. Panels show average methylation from 1,000-cell XRBS profiles for the indicated cell types. Promoters (rows, ≥25-fold combined coverage in every cell line) are grouped by the cell line in which they are specifically hypermethylated (top) or hypomethylated (bottom). Hypomethylated promoters specific to K562 cells are downsampled for visualization. A full list of differentially methylated promoters is provided in Supplementary Table 2.
B) Heatmap depicts 8kb regions (rows, n=15,202 regions) centered on H3K27ac peaks identified in K562 and OCI-AML3 ChIP-seq datasets. Rows are ordered by DNA methylation difference between both cell lines. Panels show average methylation from 1,000-cell XRBS profiles and H3K27ac signals for K562 and OCI-AML3. Cell line-specific DNA hypomethylation correlates with H3K27ac signal. Peaks not specifically hypomethylated in either cell line (‘Others’) were downsampled for visualization.
C) Heatmap depicts 2kb regions (rows, n=7,629 regions) centered on CTCF peaks identified in HL-60 and K562 CTCF ChIP-seq datasets. Rows are ordered by DNA methylation difference between both cell lines. CTCF peaks with cell type-specific DNA hypermethylation are depleted for CTCF binding in the respective cell line. Peaks not specifically hypermethylated in either cell line (‘Others’) were downsampled for visualization.