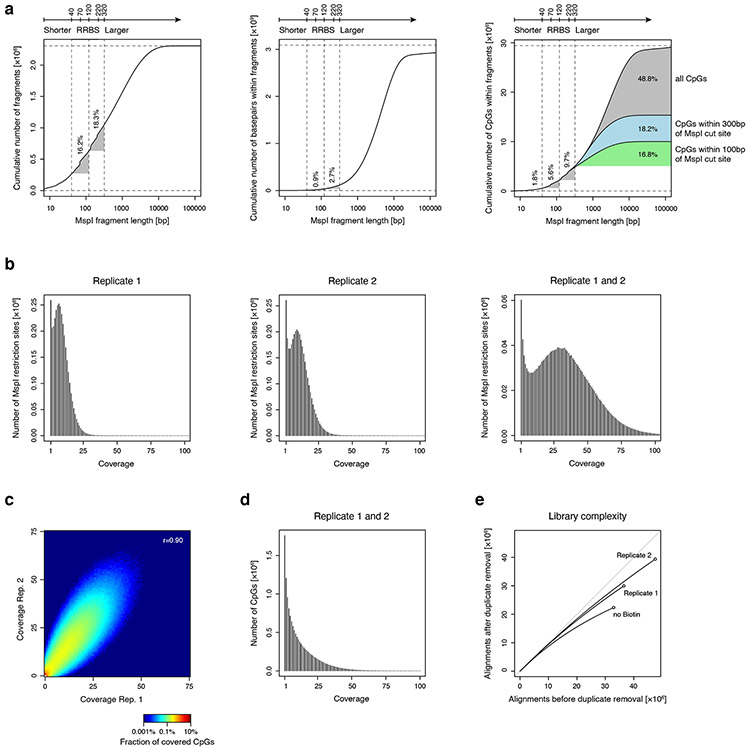
Extended Data Fig. 1 ∣. Evaluation of single MspI anchor design for methyl-CpG profiling.
A) Plots show results from an in silico MspI restriction digest analysis of the human genome. The cumulative number of MspI fragments (total of 2.3 million, left), of basepairs (total of 3.1 billion, middle), and of CpGs (total of 29.4 million, right) is shown relative to increasing MspI fragment length. Vertical dotted lines show the size range of fragments captured in typical RRBS experiments. This analysis shows that RRBS of MspI fragments 40-120 bases in length covers only 0.9% of the genome, but enriches for 5.6% of genomic CpGs. Recent implementations of RRBS (e.g. enhanced RRBS; 14,15 that consider fragments up to 320 bases in length cover an additional 9.7% of CpGs. Approximately 35.0% of CpGs that are located within 300 bases of a single MspI site are not captured by these techniques.
B) Histogram shows coverage depth of MspI restriction sites for individual replicates of a 10ng XRBS library (left, middle), and both replicates combined (right).
C) Heatmap shows coverage depth of CpGs between replicates of a 10ng XRBS library (Pearson’s r= 0.90).
D) Histogram shows coverage depth of CpGs in the combined dataset of both replicates (n=2 independently generated libraries).
E) Plot shows unique reads as a function of aligned reads in millions. With greater sequencing depth the fraction of unique reads decreases, as the chance of sampling a non-unique read (i.e. PCR duplicate) increases.