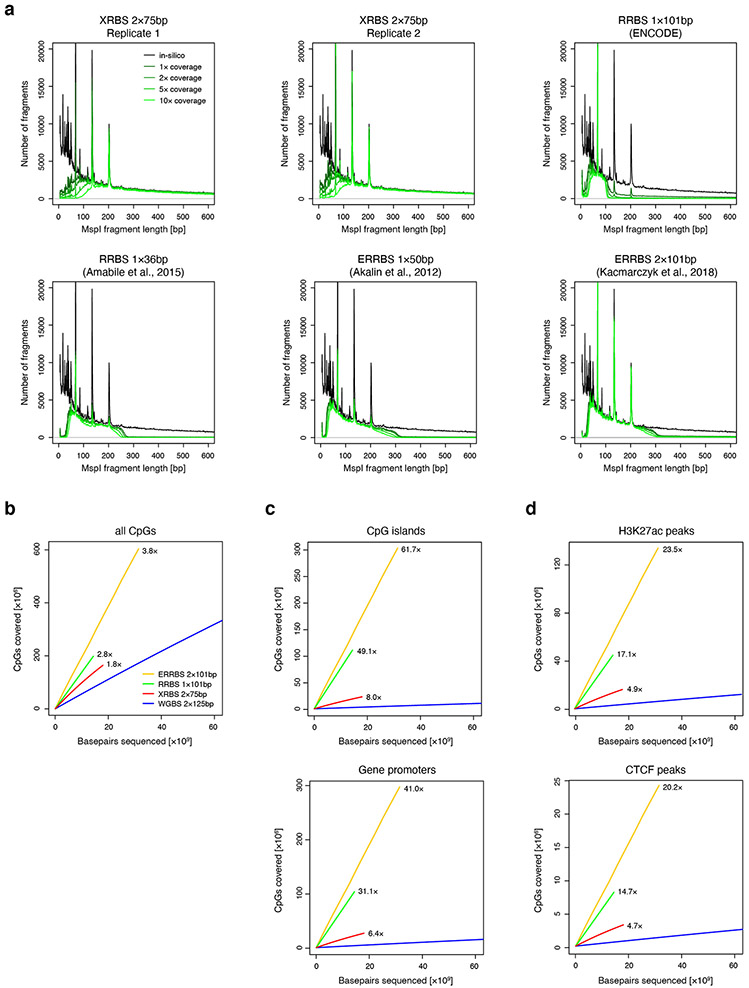
Extended Data Fig. 2 ∣. Comparison of MspI fragment detection and CpG coverage over regulatory elements.
A) Plot shows number of detected fragments plotted as a function of calculated MspI fragment length from XRBS 10ng library replicates and from public RRBS and enhanced RRBS (ERRBS) datasets. Because of the random hexamer-primed second strand elongation step, XRBS efficiently detects fragments that exceed the selected fragment size range in RRBS (ENCODE; Amabile et al.: 40-220bp) and ERRBS (70-320bp). XRBS less efficiently captures short fragments (<70bp) compared to ERRBS and RRBS. Peaks in the graph correspond to three fragments commonly generated from Alu repetitive elements.
B) Plot compares CpG coverage as a function of sequencing depth (x-axis) for XRBS (red), WGBS (blue, ENCODE), ERRBS (orange; 52) and RRBS (green, ENCODE).
C) Downsampling analysis plot as in panel B but restricted to CpGs within CpG islands (top) and gene promoters (bottom).
D) Downsampling analysis plot as in panel B but restricted to CpGs within H3K27ac peaks (top) and CTCF binding sites (bottom).