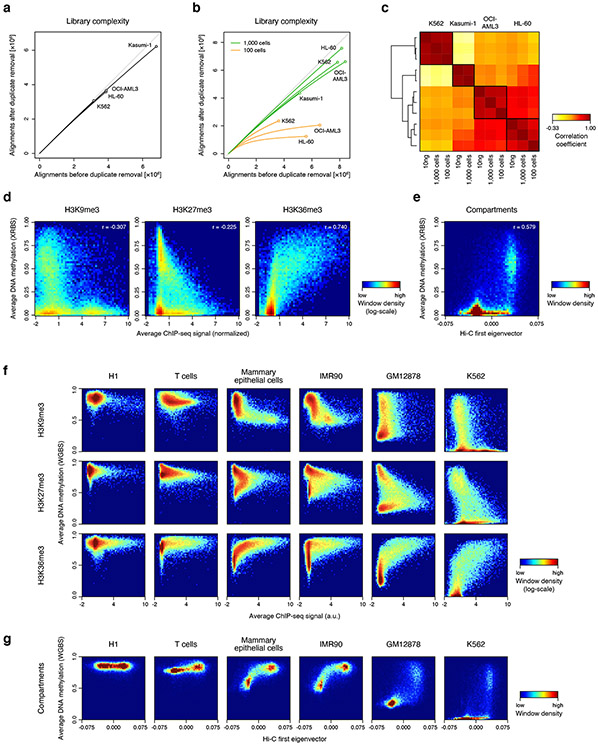
Extended Data Fig. 4 ∣. Correlation of DNA methylation with histone marks and compartment calls.
A) Plot shows unique reads as a function of aligned reads in low-coverage XRBS libraries from K562, HL-60, OCI-AML3, and Kasumi-1 cells.
B) Plot shows unique reads as a function of aligned reads in low-coverage libraries from K562, Kasumi-1, HL-60, OCI-AML3 cells. Libraries were generated from 1,000 (green) and 100 (orange) cells sorted directly into lysis buffer. Libraries generated from 1,000 cells are comparable to libraries generated from 10ng of purified DNA (panel A), whereas 100 cell libraries show reduced complexity.
C) Heatmap shows Pearson correlation of XRBS methylation profiles of 100kb windows generated from 10ng gDNA, 1,000 or 100 sorted cells across four cell lines. Dendrogram derived from unsupervised clustering is indicated to the left. Sample grouping by DNA methylation is consistent with cell identity, indicating low technical variability between input material.
D) Heatmaps show correlation between average DNA methylation values and signal for H3K9me3 (left), H3K27me3 (center), and H3K36me3 (right) in 100kb-windows for K562 cells.
E) Heatmap shows correlation between DNA methylation and the Hi-C-derived first eigenvector indicating compartment A (positive values) and compartment B (negative values) in 100kb-windows for K562 cells.
F) Heatmap shows correlation between average DNA methylation values and ChIP seq signal for H3K9me3 (top), H3K27me3 (middle), and H3K38me3 (bottom) in 100kb-windows for human H1 embryonic stem cells, primary T cells and mammary epithelial cells, and cultured IMR90, GM12878 and K562 cells.
G) Heatmap as in panel F, but shows correlation between average DNA methylation values and the Hi-C-derived first eigenvector (x-axis). Positive values correspond to compartment A and negative values correspond to compartment B. While hypomethylation of compartment B is most pronounced in K562 cells, a similar trend is also observed in other cultured cell lines and in primary mammary epithelial cells.