FIGURE 1:
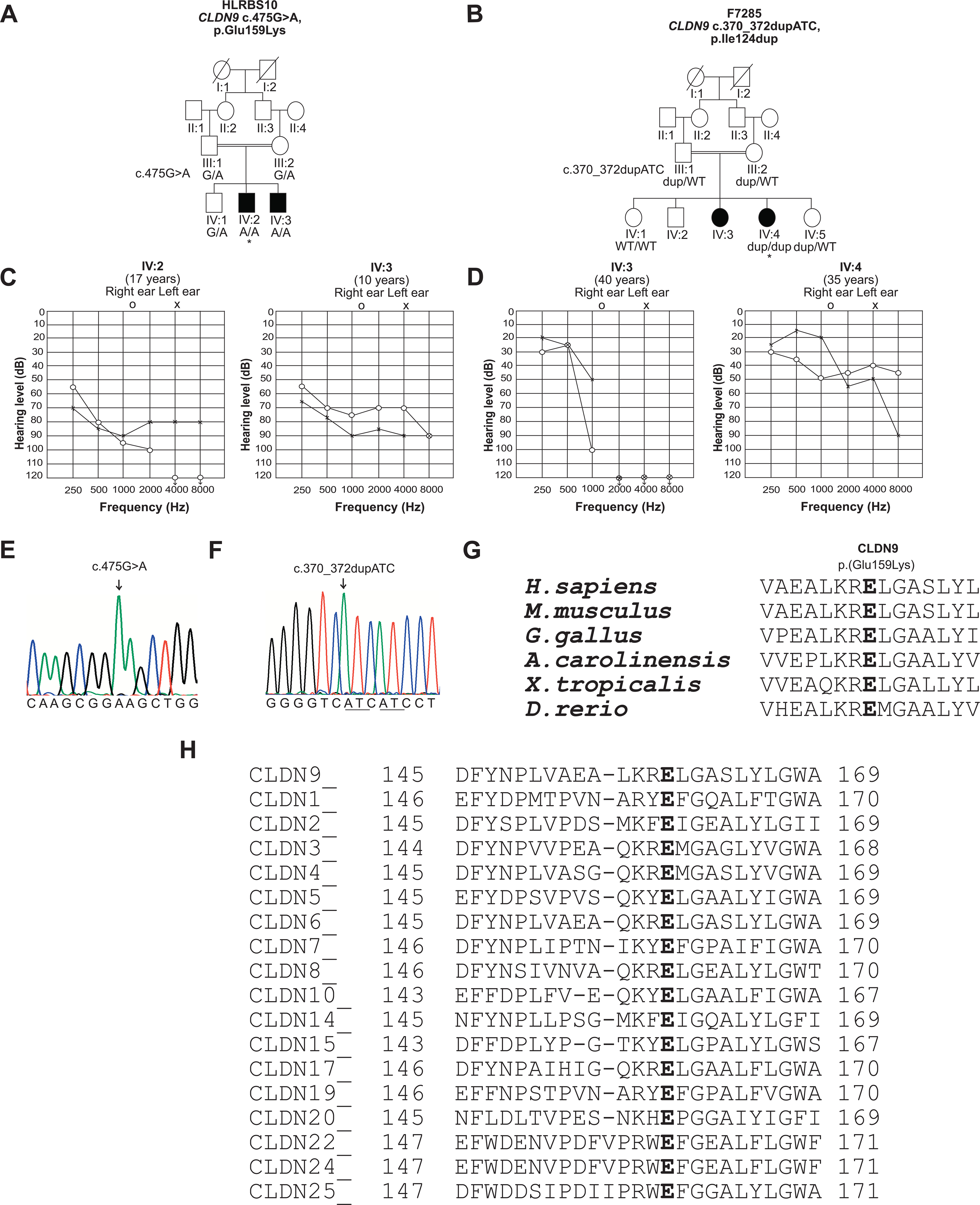
Pedigrees of families HLRBS10 and F7285, pure tone audiometry and amino acid conservation. A, Pedigree of family HLRBS10 showing segregation of the c.475G>A, p.(Glu159Lys) with hearing loss. Open symbols represent unaffected individuals, while filled circles and squares denote affected individuals. * indicate that individual IV:2 of family HLRBS10 was selected for exome sequencing. B, Pedigree of family F7285. * indicate the individual selected for exome sequencing. The c.370_372dupATC variant segregated with the phenotype. C, Audiograms of individuals IV:2 and IV:3 show asymmetric moderate to profound and moderate to severe hearing loss, respectively, in two individuals from family HLRBS10. D, In family F7285, individuals IV:3 and IV:4 have down-sloping audiograms and display an asymmetric mild hearing loss at the lower frequencies and profound deafness at the higher frequencies. E, F, Partial sequence traces indicating variants c.475G>A and c.370_372dupATC in affected individuals from the families HLRBS10 and F7285, respectively. Arrows indicate positions of the mutated residues. G, H, CLUSTALO multiple alignment shows that the glutamic acid-159 residue (Glu-159) is highly conserved in CLDN9 proteins from a variety of vertebrate species. Partial sequence alignment for human CLDN9 with its paralogues also indicates conservation of Glu-159. CLDN14, which is another tight junction protein required for normal hearing, also has a glutamate residue at the corresponding position.
