Fig. 1.
Accessible SARS-CoV-2 detection through molecular nanostructures and automated microfluidics.
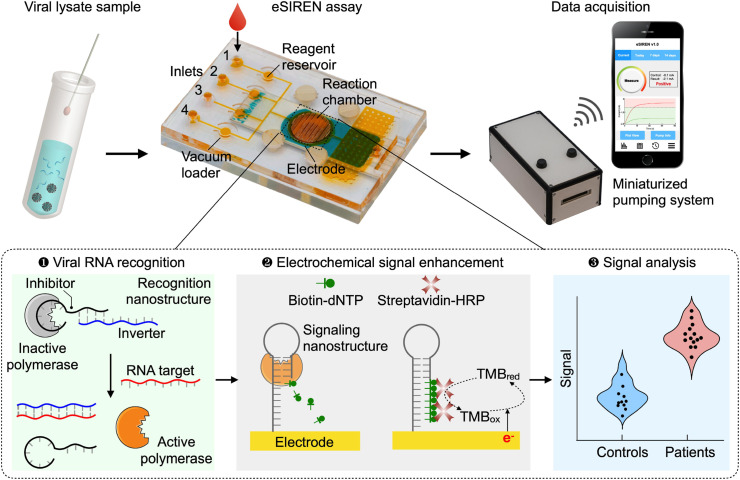
Schematics of the eSIREN platform. The platform incorporates molecular nanostructures, integrated microfluidics and a miniaturized pumping system for the direct and automated detection of viral RNA (S gene target of SARS-CoV-2). It leverages a molecular circuitry comprising catalytic enzyme-DNA nanostructures to directly recognize target RNA sequences and automated microfluidics to interface the molecular circuitry with the embedded electrodes, so as to transduce the direct target recognition into an amplified electrical signal. For the molecular circuitry, molecular nanostructures are organized to achieve three functional steps. In the target recognition step, the recognition nanostructure – a hybrid molecular complex comprises DNA strands (inhibitor and inverter sequences) that bind to and inhibit a DNA polymerase enzyme – is mixed with a clinical sample. Only in the presence of target RNA, upon target hybridization with the inverter sequence, the nanostructure dissociates to liberate strong polymerase activity. In the signal enhancement step, the activated polymerase elongates the signaling nanostructure – a self-primed DNA nanostructure immobilized onto the electrode – to incorporate biotin-modified deoxynucleotide triphosphates (biotin–dNTPs). This incorporation recruits streptavidin-conjugated horseradish peroxidase (HRP) near the electrode surface to enhance electrochemical signal amplification. The resultant electrochemical current changes are used to measure viral RNA targets. The entire assay is automated and can be completed in <20 min at room temperature, to achieve direct and user-friendly detection of SARS-CoV-2 infection near patients.