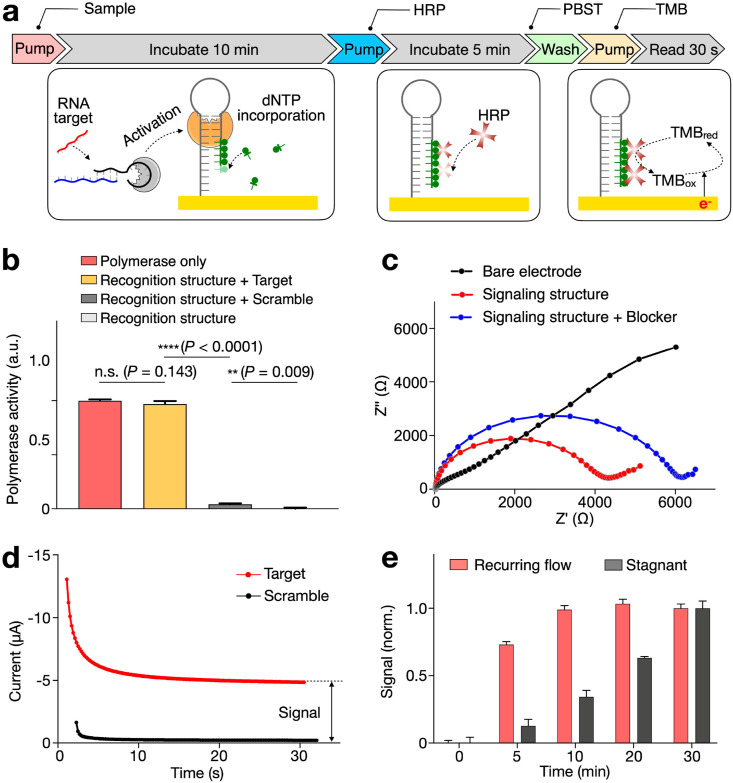
Fig. 2.
Characterization of the eSIREN assay.
(a) Workflow of the assay operation. Clinical sample, recognition nanostructure and biotin-dNTPs were first applied to the electrode surface. This was followed by a 10-min incubation, during which the recognition nanostructure was activated, in the presence target RNA, to elongate the signaling nanostructure. After a brief flushing, streptavidin-HRP was introduced for a 5-min incubation, to enable the binding of HRP onto the signaling nanostructures through streptavidin-biotin interaction. After washing with PBST buffer, redox substrate (Tetramethylbenzidine, TMB) was introduced to start the electrochemical reaction. Continuous electrochemical data acquisition was performed through a potentiostat. All assay reactions were completed at room temperature. (b) Characterization of the recognition nanostructures. Only in the presence of target RNA, the recognition nanostructures were activated. The resultant activity matched that of pure polymerase, suggesting the high specificity of the recognition nanostructures. (c) Characterization of the signaling nanostructures. Through electrochemical impedance spectroscopy, the increasing electrode resistance confirmed that the signaling nanostructures and protein blockers were successfully deposited in sequence onto the electrode surface. (d) An example of eSIREN measurements with target and control sequences. The complementary target generated a larger stabilized current, while the scrambled control generated negligible signal. (e) Incubation optimization with recurring flow. The incubation duration could be reduced by at least 3 × through the recurrent introduction of fresh reagents. All measurements were performed in triplicate and the data are presented as mean ± s.d in b, e.