FIG. 8.
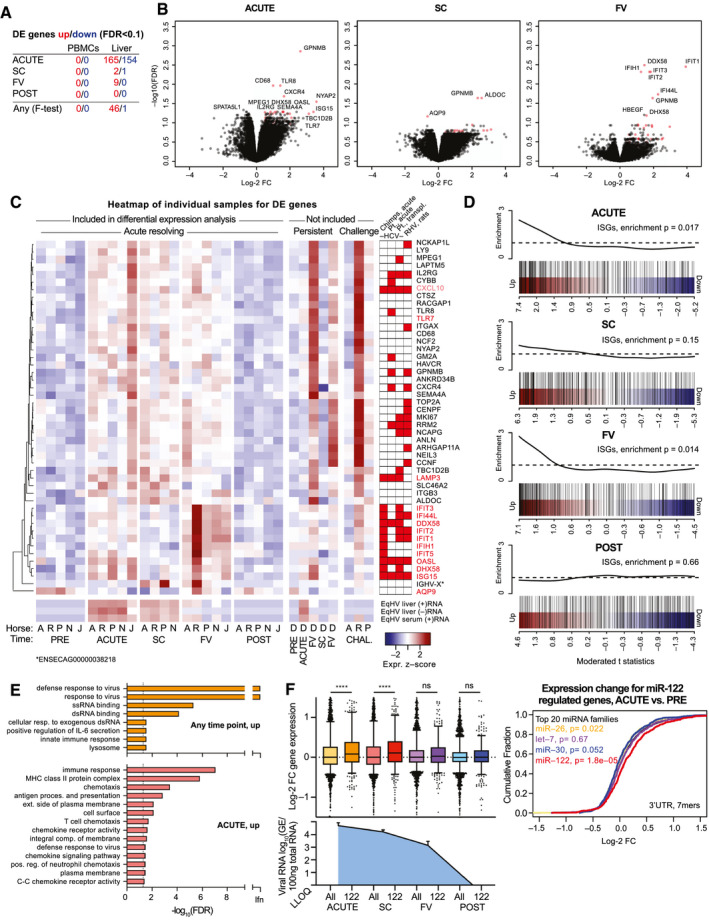
Liver transcriptional analysis reveals an ISG response. (A) The number of DE genes compared to PRE is indicated for individual time points and for multiple comparison (any). (B) Volcano plots of indicated time points compared to PRE in liver. The most significant genes are labeled. DE genes at any time point (F‐test) are indicated in red. (C) Heatmap showing scaled expression of liver DE genes at any time point across individual samples. EqHV (+/–) strand were significantly regulated. Scaled serum RT‐qPCR‐derived (+)RNA levels are shown for comparison. Genes significantly up‐regulated during HCV or RHV infection are indicated.( 17 , 18 , 19 , 29 ) (D) Gene set enrichment testing barcode plots for ISGs in the liver at time points as indicated. ROAST P values are shown. (E) GO analysis for up‐regulated genes in the liver at time points indicated. (F) Left: box plots of log‐2 FC liver gene expression for mRNAs with 7‐ or 8‐mer seed sites of any top 20 expressed miRNA (All) or miR‐122 specifically (122) in the 3′UTR is shown for time points indicated. Liver viral load is shown for comparison. Right: cumulative density function (CDF) of the log‐2 FC in liver gene expression for the ACUTE time point. mRNAs are grouped by presence of miRNA 7‐ or 8‐mer seed site in the 3′UTR. ****P < 0.0001. Data are deposited in GEO with accession number GSE158753. Abbreviations: ALDOC, aldolase, fructose‐bisphosphate C; ANKRD34B, ankyrin repeat domain 34B; ANLN, anillin actin binding protein; AQP9, aquaporin 9; ARHGAP11A, Rho GTPase activating protein 11A; CCNF, cyclin F; CENPF, centromere protein F; CTSZ, cathepsin Z; CXCL10, C‐X‐C motif chemokine ligand 10; CYBB, cytochrome B‐245 beta chain; DDX58, DExD/H‐box helicase 58; DHX58, DExH‐box helicase 58; dsRNA, double‐stranded RNA; FC, fold change; FV, falling viremia; HBEGF, heparin binding EGF‐like growth factor; GEO, Gene Expression Omnibus; GM2A, GM2 ganglioside activator; HAVCR, hepatitis A virus cellular receptor 1; IFI44L, interferon‐induced protein 44‐like; IFIH1, interferon‐induced helicase C domain‐containing protein 1; IFIT1, interferon‐induced protein with tetratricopeptide repeats 1; IFIT2, interferon‐induced protein with tetratricopeptide repeats 2; IFIT3, interferon‐induced protein with tetratricopeptide repeats 3; IFIT5, interferon‐induced protein with tetratricopeptide repeats 5; IGHV‐X, immunoglobulin variable region heavy chain; ISG15, ISG15 ubiquitin‐like modifier; ITGAX, integrin subunit alpha X; ITGB3, integrin alpha‐V/beta‐3; LAMP3, lysosomal‐associated membrane protein 3; LAPTM5, lysosomal protein transmembrane 5; LLOQ, lower level of quantification; LY9, lymphocyte antigen 9; MHC, major histocompatibility complex; MKI67, marker of proliferation Ki‐67; NCAPG, non‐SMC condensin I complex subunit G; NCF2, neutrophil cytosolic factor 2; NCKAP1L, NCK‐associated protein 1‐like; NEIL3, Nei‐like DNA glycosylase 3; NYAP2, neuronal tyrosine‐phosphorylated phosphoinositide‐3‐kinase adapter 2; OASL, 2′‐5′‐oligoadenylate synthetase‐like; PRE, preinoculation; RACGAP1, Rac GTPase activating protein 1; RRM2, ribonucleotide reductase regulatory subunit M2; SC, seroconversion; SEMA4A, semaphorin 4A; SLC46A2, solute carrier family 46 member 2; SPATA5L1, spermatogenesis‐associated 5‐like 1; ssRNA, single‐stranded RNA; TBC1D2B, TBC1 domain family member 2B; TLR7, Toll‐like receptor 7; TLR8, Toll‐like receptor 8; TOP2A, DNA topoisomerase II alpha.
