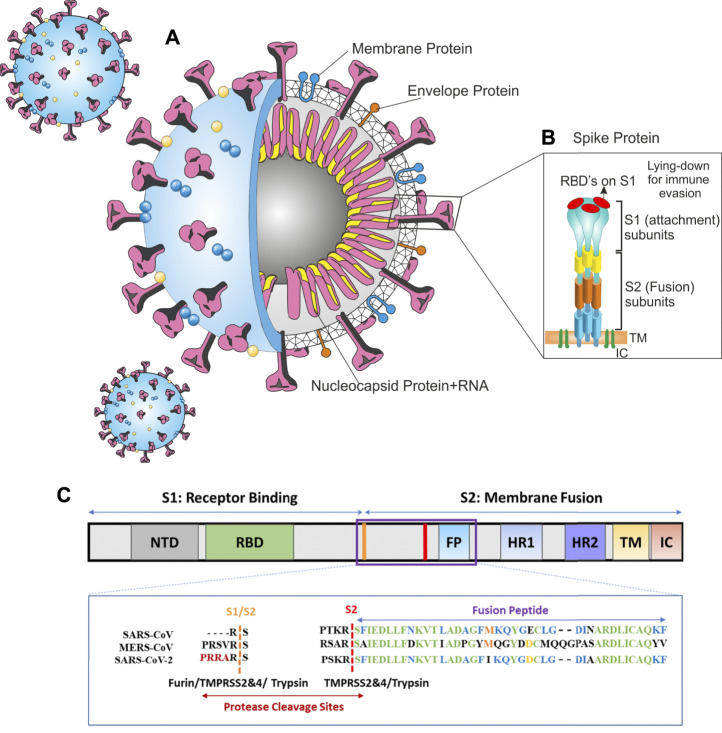
FIGURE 1.
Architecture of SARS-CoV-2 virus (A) SARS-CoV-2 schematic (B) spike protein, (C) alignment of SARS-CoV-2, MERS-CoV, and SARS-CoV sequences in protease cleavage sites S1/S2 and S2. A furin cleavage motif (RRAR) only exists in the SARS-CoV-2 spike. Trypsin, furin, and TMPRSS2 and 4 can cleave the S1/S2 site within the receptor binding domain (RBD) of S protein generating the optimal conformation for viral binding to the host ACE2 receptor. The viral membrane fusion with the host membrane S2 domain can be cleaved by TMPRSS 2 and 4. Panel C is adapted from Luan et al. (2020).