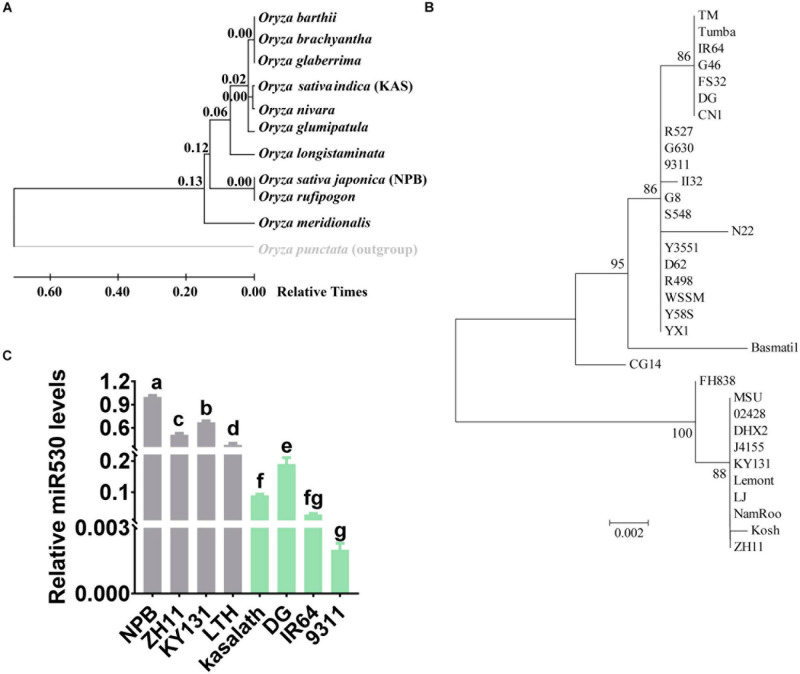
FIGURE 7.
MIR530 was diverse in rice species. (A) Phylogenetic tree inferred by analyzing the sequences of MIR530 gene (miR530 precursor with 1,000-bp upstream promoter sequence) in 11 wild rice species using the neighbor-joining method. The numbers at the nodes are relative divergence times. (B) Phylogenetic tree inferred by analyzing the MIR530 gene in 32 O. sativa accessions covering all known subpopulations and an Oryza glaberrima accession. The numbers at the nodes are relative divergence times. (C) The relative amount of mature miR530 in the indicated O. sativa japonica and O. sativa indica accessions. The RT was carried out with total RNA and miR530-specific stem-loop RT primer (Supplementary Table 1). The RT product was subsequently used as a template for qPCR to detect the amounts of miR530. The amounts of snRNA U6 were detected and used as an internal reference.