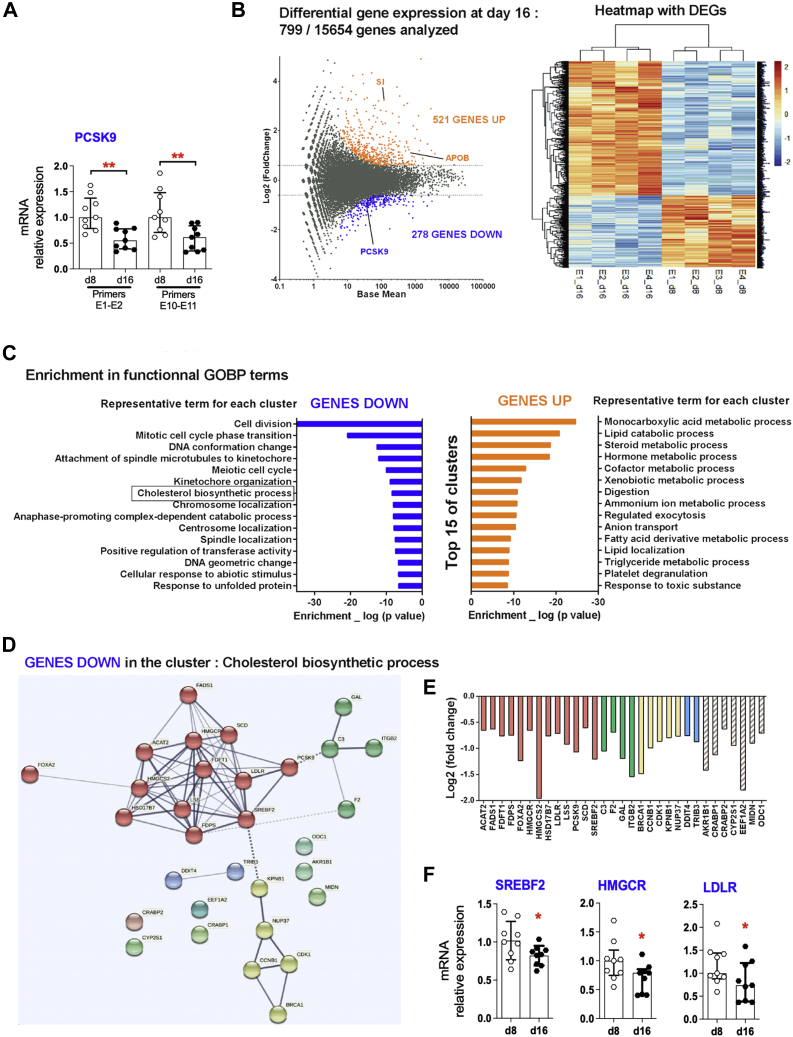
Fig. 3.
SREBP2-regulated genes, including its own gene SREBF2 and PCSK9, are downregulated in mature Caco-2 cells. Caco2 cells were recovered at d8 and at d16 after a 2-h secretion assay, and mRNAs were extracted. A: PCSK9 expression relative to that of cyclophilin was analyzed by RT-qPCR in samples from 9 independent experiments. Scatter dot plots + bars (median ± interquartile) are presented. ∗∗P< 0.01. B: 3′ Digital gene expression RNA sequencing was performed in samples from 4 independent experiments (E1 to E4) at d8 and d16. MA plot is shown in the left panel: dots corresponding to genes that are significantly upregulated at day 16 (GENES UP), downregulated at day 16 (GENES DOWN), or unchanged are colored in orange, blue, and gray, respectively. A heat map with differentially expressed genes (DEGs) is shown in the right panel: each column represents one sample showing the intensity of the expression profile per gene. C: Bar chart of functional GOBP (gene ontology biological process) terms enrichment analysis for GENES UP and for GENES DOWN is shown. D: Protein-protein interactions for the downregulated genes belonging to the cluster “Cholesterol biosynthetic process” are shown. Line thickness indicates the strength of data support. Four clusters with connected nodes are distinguished according to color of bubbles. E: Log2(fold change) for each downregulated gene of the cluster “Cholesterol biosynthetic process” is indicated. F, SREBF2, HMGCR, and LDLR expression relative to that of cyclophilin was analyzed by RT-qPCR in samples from nine independent experiments. Scatter dot plots + bars (median ± interquartile) are presented. ∗P< 0.05. HMGCR, HMG-COA reductase; LDLR, LDL receptor; PCSK9, proprotein convertase subtilisin/kexin type 9.