Abstract
The dysregulation of Notch signaling is found in many cancers and is closely related to cancer progression. As an important Notch receptor, abnormal Notch4 expression affects several tumor-cell behaviors, including stemness, the epithelial–mesenchymal transition, radio/chemoresistance and angiogenesis. In order to inhibit the oncogenic effects of Notch4 activation, several methods for targeting Notch4 signaling have been proposed. In this review, we summarize the known molecular mechanisms through which Notch4 affects cancer progression. Finally, we discuss potential Notch4-targeting therapeutic strategies as a reference for future research.
Keywords: Notch4 signaling, cancer, cancer stem cell, epithelial–mesenchymal transition, drug resistance, molecular mechanism, therapeutic perspective
Introduction
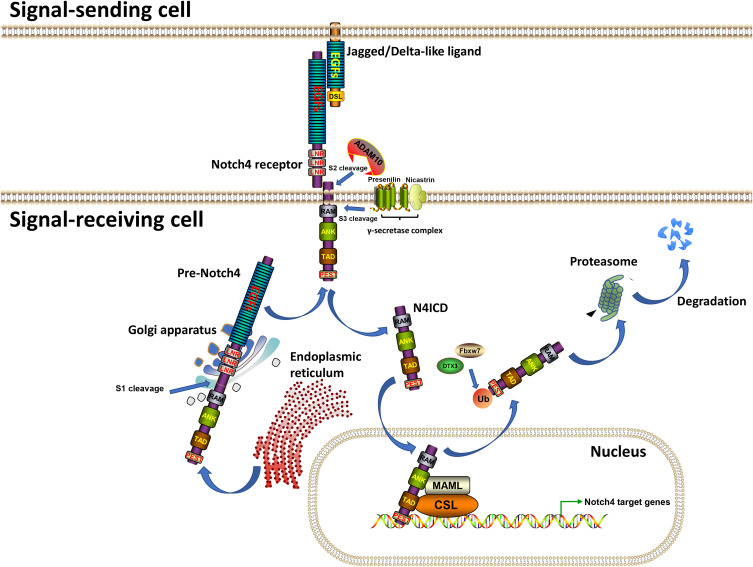
The evolutionarily conserved Notch signaling pathway is a conduit for critical cell-cell communication during cell-fate determination, regulating cell proliferation, differentiation and apoptosis.1 Four mammalian Notch receptors (Notch1-4) and five Notch ligands (Delta-like ligand (DLL) 1, 3, 4 and Jagged (JAG1) 1, 2) have been identified to date. Before being activated, Notch receptors are proteolytically cleaved three times (Figure 1). The immature transmembrane heterodimeric Notch protein (Pre-Notch) undergoes S1 cleavage in the Golgi apparatus and is subsequently trafficked to the cell surface as a mature heterodimeric transmembrane protein. On the surface of a signal-receiving cell, the epidermal growth factor (EGF) repeats of a Notch receptor interact with the EGF repeats of a Notch ligand on the surface of a neighboring signal-receiving cell, leading to S2 cleavage (mediated by A Disintegrin And Metalloprotease domain 10 (ADAM10)) and S3 cleavage (mediated by the γ-secretase complex, mainly Presenilin and Nicastrin) of the Notch receptor, which leads to the release of the Notch intracellular domain (NICD).1–3 Subsequently, NICD undergoes nuclear translocation and interacts with the effector transcription factor CSL. Transcription co-activators such as MAML are then recruited to the NICD-CSL complex, resulting in transcriptional activation of downstream target genes. Finally, NICD undergoes proteasomal degradation induced by E3 ubiquitin-ligases such as FBXW7 and DTX3.1–5
Figure 1.
The transduction process of Notch4 signaling. Note: Generated using ScienceSlides graphics from Visiscience.
Abbreviations: EGFs, Epidermal Growth Factor repeats; LNRs, Lin-Notch repeats; RAM, RBPJ-association module; ANK, Ankyrin; TAD, Trans-activation domain; PEST, proline (P), glutamic acid (E), serine (S), threonine (T)-rich; DSL, Delta/Serrate/LAG-2; ADAM10, A Disintegrin And Metalloprotease domain 10; MAML, Mastermind-like protein; CSL, CBF-1 (RBPJ)/suppressor of hairless/Lag1; Fbxw7, F-box and WD repeat domain containing 7; DTX3, Deltex E3 ubiquitin ligase 3; Ub, Ubiquitin.
Aberrant Notch signaling has been discovered in many cancer types, and it is known to play unique roles during cancer occurrence and progression.6–10 The gene encoding Notch4 protein is located on chromosome 6p21.32 (6: 32194843–32224067), and contains 31 exons. Mutations and amplifications of the Notch4 gene, which partly lead to gain-of-function of Notch4 protein, have been frequently reported in human cancers.11–19 To date, numerous studies have explored the roles of Notch4 in cancer, and especially its related molecular mechanisms. Most studies suggested that Notch4 is abnormally overexpressed during cancer development and is involved in the regulation of several tumor-cell behaviors, mainly in stem cell-like self-renewal, epithelial–mesenchymal transition (EMT), radio-/chemo resistance and angiogenesis (See details in the following parts). Here, we systematically reviewed the expression and clinical significance of Notch4 in different cancers, as well as the underlying molecular mechanisms that mediate the related tumor-cell behaviors. Additionally, we propose potential therapeutic strategies to inhibit Notch4 expression and/or block Notch4 signal transduction. Overall, this review provides a comprehensive overview of the functions of Notch4 in cancer and discusses potential future directions in the research on Notch4-targeting therapeutic approaches.
Expression of Notch4 and Its Clinicopathologic and Prognostic Significance in Cancer
As shown in Table 1, the expression of Notch4 and its clinicopathologic and prognostic significance in cancer have been widely reported. Notch4 overexpression was found in hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma, melanoma, colorectal carcinoma (CRC), oral squamous cell carcinoma (OSCC), breast carcinoma (BC), gastric carcinoma, non-small cell lung carcinoma (NSCLC) and acute myeloid leukemia (AML). In all these cancers, it is associated with a series of clinicopathologic features such as high pathological grade, advanced tumor stage and metastasis, as well as poor prognosis in terms of OS and DFS.20–31 However, low expression of Notch4 was found in endometrial carcinoma and CRC tissues.32–34 In two studies encompassing 256 cases of CRC and 1648 cases of ovarian carcinoma (OC), low Notch4 expression was significantly correlated with poor OS of cancer patients.34,35 These findings indicate that the expression levels and prognostic value of Notch4 in different types of cancer may be different and need further exploration.
Table 1.
The Clinicopathologic and Prognostic Significance of Notch4 in Cancer
| Cancer Type | Sample Count | Clinicopathologic and Prognostic Significance | Reference |
|---|---|---|---|
| Hepatocellular carcinoma | 288 | High Notch4 expression correlates with low edmondson grade, low | [20] |
| AJCC T-stage, no microvascular invasion, no intrahepatic metastasis, and low serum AFP level. | |||
| Intrahepatic cholangiocarcinoma | 41 | High Notch4 expression predicts poor OS. | [21] |
| High Notch4 expression correlates with high serum CA125 level. | |||
| Melanoma | 120 | High Notch4 expression predicts poor OS. | [22] |
| High Notch4 expression correlates with high AJCC T-stage, and lymphatic and/or distant metastasis. | |||
| Glioblastoma | 70 | Correlate with low microvessel density. | [64] |
| Colorectal carcinoma | 248 | High Notch4 expression predicts poor OS and DFS. | [28] |
| High Notch4 expression correlates with advanced N stage, M stage, lymphovascular invasion, and CEA status. | |||
| 290 | High Notch4 expression predicts poor OS. | [29] | |
| 256 | Low Notch4 expression predicts poor OS and DFS. | [34] | |
| Low Notch4 expression correlates with poor differentiation, advanced | |||
| TNM stage, deep invasion, and lymph node metastasis. | |||
| Oral squamous cell carcinoma | 60 | High Notch4 expression correlates with poor differentiation, advanced clinical stage, perineural invasion, and lymph node metastasis. | [23] |
| Ovarian carcinoma | 1648 | Low Notch4 expression predicts poor OS. | [35] |
| Breast carcinoma | 98 | High Notch4 expression predicts poor OS. | [24] |
| High Notch4 expression correlates with low ER and PR status, large tumor size, advanced TNM stage, and lymph node metastasis. | |||
| 180 | High Notch4 expression correlates with high ER and PR status. | [25] | |
| 594 | High Notch4 expression predicts poor OS. | [26] | |
| TNBC | 68 | High Notch4 expression predicts poor OS. | [26] |
| Gastric carcinoma | 876 | High Notch4 expression predicts poor OS. | [27] |
| Non-small cell lung carcinoma | 335 | High Notch4 expression predicts poor OS. | [86] |
| 189 | High Notch4 expression predicts poor OS. High Notch4 expression correlates with large tumor size, advanced TNM stage, and distant/lymph node metastasis. |
[30] | |
| Acute myeloid leukemia | 79 | High Notch4 expression predicts poor OS. | [31] |
Notch4 Signaling and Cancer Stem Cell Properties
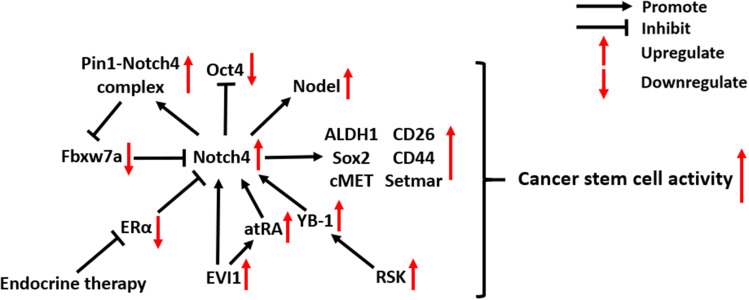
Notch4 is considered a novel biomarker of cancer stem cells (CSCs) - undifferentiated cancer cells with high tumorigenic potency. In BC, Notch4 activity is substantially enriched in ESA+/CD44+/CD24− CSCs, and Notch4+ tumor cells show CSC-like behavior, including self-renewal and lineage differentiation.26,36,37 Notch4 activation in breast CSCs is also critical for the expression of another CSC biomarker, ALDH1, whose activity can be inhibited by Notch4 knockdown.38 The high expression of Notch4 in CSCs was also discovered in microarray and bioinformatic analyses of melanoma. Subsequent in vivo experiments confirmed that Notch4+ CSCs facilitate the EMT, invasion and metastasis of melanoma.22 In addition, Notch4 plays an important role in colorectal CSCs by regulating transcription factors related to stemness. Notch4 inhibition in colorectal CSCs led to an increase of Oct4 expression but also significantly decreased the expression of Sox2, cMET, CD26, CD44 and SETMAR.39
Several mechanisms are associated with Notch4 signaling in CSCs (Figure 2). In ecotropic virus integration site 1 (EVI1)-positive AML, a particularly aggressive subtype, Notch4 is directly targeted and activated by EVI1 and its downstream factor all-trans retinoic acid (atRA), which maintains cancer cell stemness.40 In breast CSCs, Notch4 can escape from proteasomal degradation induced by ubiquitin-ligase Fbxw7a due to its interaction with the peptidyl-prolyl isomerase Pin1.41 Y-box binding protein-1 (YB-1), an oncogenic transcription factor activated by the p90 ribosomal S6 kinases (RSK), can upregulate Notch4 signaling in breast CSCs.42 Blocking RSK/YB-1 signaling can effectively prevent Notch4 activation, which reverses the undifferentiated state of breast CSCs.42 In addition, Notch4 activity in breast CSCs in response to endocrine therapy was found to be closely related to estrogen receptor alpha (ERα), which is discussed in part 5.1.
Figure 2.
Molecular mechanisms of Notch4 signaling in cancer stem cells.
Abbreviations: Pin1, Peptidylprolyl cis/trans isomerase, NIMA-interacting 1; Oct4, Organic cation/carnitine transporter4; Fbxw7a, F-box and WD repeat domain containing 7a; ALDH1, Aldehyde dehydrogenase7; Sox2, SRY-box transcription factor 2; ERα, Estrogen receptor alpha; EVI1, Ecotropic virus integration site 1; atRA, all-trans retinoic acid; YB-1, Y-box binding protein-1; RSK, Ribosomal S6 kinase.
Notch4 Signaling in Cancer EMT and Metastasis
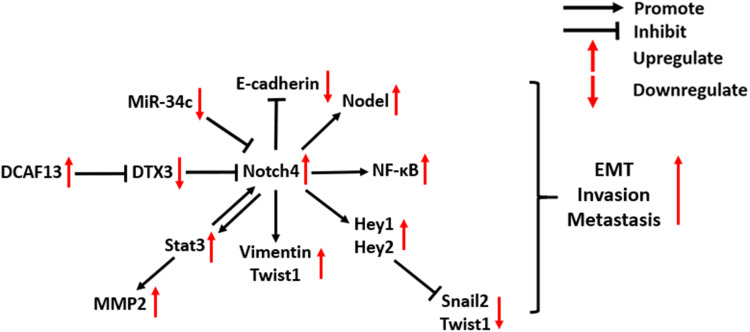
High expression of Notch4 was found to be associated with tumor metastasis in clinical studies of several types of cancer, including salivary adenoid cystic carcinoma,43 OSCC,23 melanoma,44 BC,24 and NSCLC.30 As shown in Figure 3, Notch4 signaling is involved in several molecular mechanisms of cancer EMT and metastasis. In prostate carcinoma (PC), Notch4 signaling promotes the growth, metastasis and EMT of tumor cells, which is dependent on the activation of NF-κB signaling.45 In melanoma and head and neck squamous cell carcinoma, Notch4 signaling triggers the EMT by increasing the expression of EMT markers such as Vimentin and Twist1, as well as decreasing E-cadherin expression.22,46 In addition, EMT-promoting Nodal is a downstream target of Notch4 signaling in aggressive types of melanoma. The inhibition of Notch4/Nodal axis diminishes clonogenicity and impairs the vasculogenic mimicry of melanoma.47,48 However, Menon et al found that Notch4 signaling also has an anti-EMT role in melanoma, which relies on non-canonical Notch signaling.49 The activation of Hey1 and Hey2, which are the downstream targets of Notch4 signaling, can suppress Snail2 and Twist1 promoter activity by direct binding, thereby inhibiting the EMT of melanoma.49 These findings illustrate the multifaceted roles of Notch4 in regulating the metastasis of melanoma.
Figure 3.
Molecular mechanisms of Notch4 signaling in cancer EMT and metastasis.
Abbreviations: DCAF13, DDB1 and CUL4 associated factor 13; DTX3, Deltex E3 ubiquitin ligase 3; Stat3, Signal transducer and activator of transcription 3; MMP2, Matrix metallopeptidase 2; NF-κB, Nuclear factor kappa B; EMT, Epithelial–mesenchymal transition.
Notch4 also plays a critical role in promoting BC metastasis. A recent study reported that Notch4 overexpression promotes BC cell invasion, which is induced by the RNA-binding protein (RBP) DCAF13, which inhibits the activation of the E3 ubiquitin-protein ligase DTX3 and prevents Notch4 degradation.5 An in vitro experiment indicates that there is a positive crosstalk between Notch4 and EMT-promoting Stat3-MMP signaling in BC metastasis: N4ICD can physically interact with the EMT-promoting Stat3 protein to activate it in BC cells, which leads to the upregulation of MMP2 expression. Stat3 signaling, in turn, maintains the activation of Notch4 and N4ICD.50 In a mouse model of BC metastasis into the liver, inhibition of Notch4 signaling by a γ-secretase inhibitor, DAPT, reduced the micrometastatic tumor burden, suggesting that Notch4 is a potential therapeutic target for delaying BC metastasis.50
In breast CSCs, Notch4 signaling can transcriptionally upregulate the expression of Slug, which induces the EMT.26 In addition, the Notch4-induced EMT process of breast CSCs is also promoted by the downregulation of the tumor-suppressive miRNA miR-34c. Tumorspheres derived from cancer cells have been proven to display characteristics of CSCs. Ectopic expression of miR-34c in breast CSCs reduced Notch4 expression, reducing the formation of tumorspheres, as well as the self-renewal, migration, and EMT propensity of CSCs.51
Notch4 Signaling and Radio-/Chemo-Therapy Resistance in Cancer
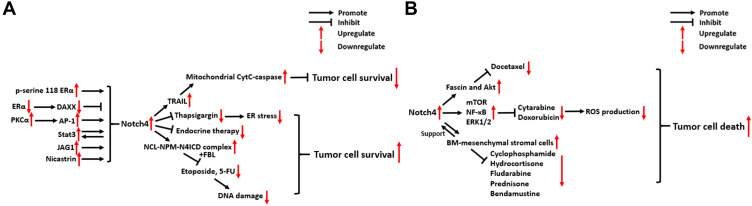
There is emerging evidence that Notch4 signaling regulates cancer radio- and chemoresistance through several different mechanisms, especially in BC (Figure 4). In this part, we mainly discuss the relationship between Notch4 signaling and radio/chemoresistance in BC and other types of cancer.
Figure 4.
Molecular mechanisms of Notch4 signaling in cancer radio/chemoresistance. (A) Breast carcinoma. (B) Other types of cancer.
Abbreviations: ERα, Estrogen receptor alpha; DAXX, Death-associated factor 6; PKCα, Protein kinase C alpha; Stat3, Signal transducer and activator of transcription 3; JAG1, Jagged 1; TRAIL, TNF-related apoptosis-inducing ligand; CytC, cytochrome c; ER, Endoplasmic Reticulum; NCL, Nucleolin; NPM, Nucleophosmin; FBL, Fibrillarin; Akt, Serine/threonine kinase 1; mTOR, mechanistic target of rapamycin kinase; NF-κB, Nuclear factor kappa B; ERK1/2, Extracellular regulated MAP kinase1/2; ROS, Reactive oxygen species.
Breast Carcinoma
In ERα+ BC, ERα activation was found to suppress the activity of Notch4+ CSCs by stabilizing the expression of Death-associated factor 6 (DAXX).52 However, this inhibitory effect is reversed in response to the inhibition of ERα by endocrine therapies, which leads to the activation of Notch4+ CSCs.52 In addition, mutations in the hormone binding domain (HBD) of ERα can also activate Notch4 signaling in breast CSCs, which is dependent on ERα phosphorylation at serine 118.53 JAG1/Notch4 signaling in ALDH1+/ER− breast CSCs expands their population and stimulates their activity following tamoxifen (TAM) or fulvestrant treatment, which contributes to tumor recurrence.54 Nicastrin (NCT), a core component of the γ-secretase (GS) complex, drives Notch4 signaling to support CSC properties and induce TAM resistance, which is linked to the acquisition of a mesenchymal phenotype in BC.38,55,56 In TAM-resistant BC cells, PKCα signaling was found to selectively increase Notch4 expression by increasing the binding of activator protein-1 (AP-1) to the Notch4 promoter, thus contributing to endocrine resistance and chemoresistance.57 In addition, Notch4/Stat3 signaling was also found to contribute to the EMT of TAM-resistant BC cells (See part 3).50
Another key mechanism through which Notch4 promotes radio/chemoresistance in BC is the mitigation of genomic damage. BC cells pretreated with short interfering RNAs (si-RNAs) against Notch4 showed increased sensitivity to apoptosis following treatment with the DNA-damaging agents etoposide or 5-fluorouracil (5-FU).58 Immunostaining and co-localization analysis indicated that N4ICD forms a complex with the nucleolar proteins NCL and NPM, thereby protecting BC cells from apoptosis triggered by genomic stressors. This process also involves another nucleolar protein, FBL, and depends on the nucleolar localization of N4ICD, but not N4ICD canonical signaling in the nucleoplasm.58
In a previous study, Shambhavi et al demonstrated that Notch4 signaling can inhibit BC cell death induced by thapsigargin, a calcium pump inhibitor that induces ER stress, as well as the genotoxic agent etoposide.59 In addition, they revealed that Notch4 signaling can also intrinsically enhance the sensitivity of BC cells to TRAIL-induced apoptosis, which proceeds via the caspase-8-Bid-Bak mitochondrial cytochrome c-caspase apoptotic pathway.59
Other Types of Cancer
In pancreatic carcinoma (PC), Notch4 overexpression contributes to docetaxel resistance by upregulating fascin and Akt expression. Inhibiting Notch4 signaling re-sensitizes PC cells to the action of docetaxel.60 In B-cell acute lymphoblastic leukemia (B-ALL), Notch4 signaling inhibits the production of ROS induced by the chemotherapeutic agents cytarabine (Ara-C) and doxorubicin. Additionally, the mTOR, NF-κB and ERK1/2 signaling pathways are activated by Notch4 in the process of ROS-dependent Notch4-mediated tumor cell survival.61 In chronic lymphocytic leukemia (CLL), activated Notch4 signaling between bone marrow (BM)-mesenchymal stromal cells and CLL cells can rescue CLL cells from apoptosis following treatment with various chemotherapy drugs, including cyclophosphamide, hydrocortisone, fludarabine, prednisone and bendamustine.62 These findings illustrate the multifaceted mechanisms by which Notch4 signaling promotes drug resistance in different types of cancer.
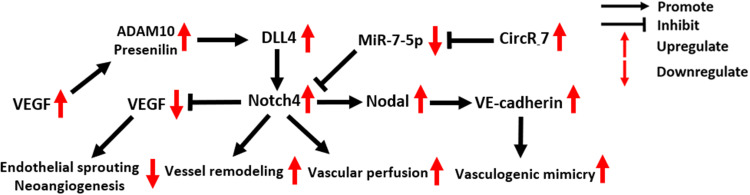
Notch4 in Tumor Vasculature
Notch4 is overexpressed in the vasculature of mouse and human mammary tumors,63,64 and it is required for tumor angiogenesis, vascular perfusion and vasculogenic mimicry (Figure 5). Notch4 expression in the endothelium has a similar effect to the expression of DLL4, one of the most important ligands that activate Notch4 in tumor vasculature, inhibiting angiogenesis by restricting endothelial sprouting and neo-angiogenesis induced by VEGF signaling.9,65–67 In an immunohistochemical study of 70 patients with primary glioblastoma, a positive correlation between vascular Notch4 and DLL4 expression was detected.64 In addition, elevated vascular expression levels of Notch4 and DLL4 were found to be associated with decreased tumor microvessel density (MVD), while VEGF expression showed an opposite trend, which indicates the negative regulatory effect of DLL4/Notch4 signaling in tumor angiogenesis.64
Figure 5.
Molecular mechanisms of Notch4 signaling in tumor vasculature.
Abbreviations: VEGF, Vascular Endothelial Growth Factor; ADAM10, A Disintegrin And Metalloprotease domain 10; DLL4, Delta-like ligand 4.
In addition to being inhibited by DLL4/Notch4 signaling, VEGF secreted by tumor cells can promote DLL4/Notch4 signaling between tumor endothelial cells (ECs) by upregulating ADAM10 and Presenilin expression.68 The activation of N4ICD then activates reverse Ephrin-B2 signaling, which subsequently induces tumor vessel remodeling, a critical step in the formation of structurally and functionally abnormal tumor vessels.68
Costa et al revealed the indispensable role of Notch4 for initial vascular perfusion in tumors.63 In blood vessels of C57BL/6 MMTV-PyMT (mouse mammary tumor virus-polyoma middle tumor-antigen) transgenic breast tumor mice, the number of perfused vessel segments was significantly decreased in tumors of Notch4−/− hosts compared to wild-type hosts.63
Vasculogenic mimicry (VM) is an epithelium-independent tumor microcirculation system that mimics the embryonic vascular network pattern to supply oxygen and nutrients for tumor cells.69 In two clinical studies of 189 NSCLC and 85 HCC patients, the expression of Notch4 and VM showed a significant positive correlation.30,70 Recently, the oncogenic circular RNA 7 was found to inhibit miR-7-5p expression in HCC, activating Notch4 expression and thus promoting VM formation.71 Suppressing the expression of circRNA 7 or Notch4, as well as recovering miR-7-5p expression, could effectively block VM in HCC.71 In aggressive melanoma, VM formation induced by Notch4 activity is dependent on the expression of Nodal and VE-cadherin. Accordingly, blocking Nodal/VE-cadherin signaling may be a potential strategy for inhibiting Notch4-induced VM formation in melanoma.48
Therapeutic Strategies for Targeting Notch4 in Cancer
Since Notch4 is widely considered to be a key player in oncogenesis, it may be useful to block Notch4 signaling in cancer. Here, we summarize known methods/strategies for inhibiting Notch4 expression and blocking Notch4 signal transduction.
Inhibiting Notch4 Expression and Activation
In pre-clinical experiments, one effective method to directly inhibit Notch4 expression is silencing Notch4 gene. As shown in Table 2, Notch4 knockdown using short interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) suppresses the tumorigenicity of cancer cells and delays tumor development in several types of cancer.22,23,36,38,39,43,45,46,51,60,70,72 However, no effective delivery vehicles for Notch4-specific siRNAs/shRNAs have been reported, which limits their value for clinical application.
Table 2.
Knockdown of Notch4 Suppresses Notch4 Oncogenic Signaling in Cancer
| Cancer type | Observations | Reference |
|---|---|---|
| Salivary adenoid cystic carcinoma | Silencing of Notch4 by si-RNA inhibited tumor cell invasion in vitro. | [43] |
| Breast carcinoma | Silencing of Notch4 by si-RNA inhibited the stemness, mammosphere formation, migration, and EMT of CSCs in vitro. | [22] |
| Silencing of Notch4 by si-RNA inhibited the proliferation, colony formation and invasiveness of tumor cells in vitro and inhibited the tumorigenicity and tumor volume in vivo. | [23] | |
| Silencing of Notch4 by sh-RNA reduced mammosphere colonies and tumor formation both in vivo and in intro | [36] | |
| Silencing of Notch4 by si-RNA inhibited CSC activity in vitro. | [38] | |
| Colon carcinoma | Silencing of Notch4 by si-RNA inhibited the gene expression of stemness’s markers of tumor cells in vitro. | [39] |
| Melanoma | Silencing of Notch4 by sh-RNA inhibited the invasion, migration, and VM formation of tumor cells in vitro. | [45] |
| Oral squamous cell carcinoma | Silencing of Notch4 by si-RNA inhibited the proliferation and migration of tumor cells in vitro. | [51] |
| Pancreatic carcinoma | Silencing of Notch4 by si-RNA inhibited the viability, migration, invasion and drug resistance of tumor cells in vitro. | [60] |
| Hepatocellular carcinoma | Silencing of Notch4 by si-RNA inhibited the invasion and vasculogenic mimicry formation of tumor cells in vitro. | [70] |
| Prostate carcinoma | Silencing of Notch4 by si-RNA inhibited the growth, migration, invasion and EMT of tumor cells in vitro. | [72] |
| Head and neck squamous cell carcinoma | Silencing of Notch4 by si-RNA inhibited the proliferation, EMT and drug resistance of tumor cells in vitro. | [46] |
MicroRNAs (miRNAs) are a class of short non-coding RNAs that control tumor development and progression by regulating gene expression at the post-transcriptional level.73 A total of eight miRNAs, including miR-7, miR-34a, miR-34c, miR-96, miR-183, miR-181c, miR-302 and miR-1179, were found to target Notch4 and inhibit its expression in cancer cells (Table 3).47,51,71,74–77 However, their expression levels are aberrantly downregulated in tumor tissues, which is partly caused by hypermethylation of their gene promoters, or overexpression of the upstream circRNA.51,71,76 MiRNA mimics, DNA demethylation inducers or circRNA inhibitors can recover the expression of Notch4-targeted miRNAs in cancer cells, thereby reducing Notch4 expression and inhibiting tumor progression.
Table 3.
Multiple miRNAs Inhibit Cancer Progression by Negatively Regulating Notch4
| miRNA | Cancer Type | Observations | Reference |
|---|---|---|---|
| miR-7 | Hepatocellular carcinoma | MiR-7 suppresses the vasculogenic mimicry formation of HCC by targeting Notch4. | [71] |
| miR-34a | Pancreatic Ductal Carcinoma | MiR-34a suppresses the survival, invasion and migration of tumor cells and tumor growth by targeting Notch1, 2 and 4. | [74] |
| miR-34c | Breast carcinoma | MiR-34c suppresses the self-renewal, migration and EMT process of tumor-initiating cells by targeting Notch4. | [51] |
| miR-96 and miR-183 | EBV-associated nasopharyngeal carcinoma | MiR-96 and miR-183 suppress the growth, colony formation and the stem cell-like of tumor cells by inhibiting the expression of NICD3 and NICD4. | [75] |
| miR-181c | Gastric carcinogenesis | MiR-181c suppressed tumor cell proliferation by targeting Notch4 and KRAS. | [76] |
| miR-302 | Melanoma | MiR-302 suppresses the tumorigenic phenotype of aggressive tumor cells by targeting Notch4. | [47] |
| miR-1179 | Breast carcinoma | MiR-34a suppresses the proliferation, invasion and migration of tumor cells by targeting Notch1, 4 and Hes1. | [77] |
mAbs against Notch receptors or ligands, such as anti-Notch1 antibodies, anti-DLL3 antibodies and anti-DLL4 antibodies, have shown promising effects in both pre- and clinical studies.6,9 In aggressive melanoma, the treatment of tumor cells with a goat antibody against human Notch4 was found to block Notch4 signaling, which inhibited tumor cell proliferation, impaired VM and diminished clonogenicity in vitro.48 In B-ALL and CLL, treatment with a combination of an anti-Notch4 antibody and GSI XII was able to block Notch4 signaling between BM-mesenchymal stromal cells and tumor cells, thus reducing tumor cell survival.62,78 Although the effects of anti-Notch4 antibodies in cancer treatment have been investigated in pre-clinical studies, we are not aware of any clinical studies trials investigating the potential effects of antibody drugs targeting Notch4.
Blocking Notch4 Signal Transduction
There are several potential strategies for interfering with Notch4 signal transduction. Targeting the γ-secretase complex is the most commonly used approach for blocking Notch signal transduction, and Notch4 signaling is no exception (Table 4). Gamma-secretase complex inhibitors (GSIs) have shown potent inhibitory effects on oncogenic Notch4 signaling in pre-clinical in vivo and in vitro experiments in BC and B-ALL.36,37,50,54,57,61,78 In addition, Nicastrin was recently identified as a target of the γ-secretase complex that can be used to block Notch4 signaling, and its inhibition can suppress the EMT and CSC properties of tumor cells.55,56
Table 4.
Targeting the γ-Secretase Complex Blocks Notch4 Signal Transduction in Cancer
| Cancer Type | Drug | Target | Effects | Reference |
|---|---|---|---|---|
| Breast carcinoma | DAPT (GSI-IX), Dibenzazepine and MK-0752 | γ-secretase complex | Inhibit CSC activity in vivo and reduce tumor growth in vivo by blocking Notch4 signaling. | [36] |
| MRK-003 | γ-secretase complex | Inhibit the growth of endocrine therapy-resistant tumors both in vivo and in vitro by blocking Notch4 signaling. Reduce the tumor-initiating capacity of tumor cells in vivo by blocking Notch4 signaling. |
[37, 57] | |
| RO4929097 | γ-secretase complex | Inhibit CSC activity in endocrine therapy-resistant tumors both in vivo and in vitro by blocking Notch4 signaling. | [54] | |
| DAPT | γ-secretase complex | Reduce micro-metastatic tumor burden in vivo by blocking Notch4 signaling. | [50] | |
| B-cell acute lymphoblastic leukemia | DAPT and GSI-XII | γ-secretase complex | Inhibit the survival, proliferation and chemoresistance of tumor cells both in vivo and in vitro by blocking Notch4 signaling. | [61,78] |
| Breast carcinoma | Nicastrin mAbs PF03084014, and RO4929097 | Nicastrin γ-secretase complex | Inhibit the EMT, stem cell content and properties of endocrine therapy-resistant tumor cells in vitro by blocking Notch4 signaling. | [56] |
| Nicastrin knockdown | Nicastrin | Inhibit the EMT and CSC activity in vitro and inhibit tumor formation in vivo by blocking Notch4 signaling. | [55] |
Shambhavi et al reported that knockdown of CSL or ectopic expression of dominant negative Mastermind-Like 1 (DN-MAML) prevents the formation of the N4ICD-CSL-MAML complex in the nuclei of tumor cells, inhibiting the transcriptional outcomes of Notch4 signaling. These findings indicate that the N4ICD-CSL-MAML complex is a potential target for blocking Notch4 signal transduction.59 In addition, promoting the proteasomal degradation of N4ICD in CSCs by recovering the expression of E3 ubiquitin-ligase Fbxw7a or DTX3 was also confirmed effective in blocking Notch4 signal transduction and its oncogenic function.5,41
The main challenge of the discussed treatment strategies is their potential pan-Notch inhibitory effect. GSIs are well-known pan-Notch inhibitors that show serious adverse effects in clinical trials, such as gastrointestinal toxicity.79 In addition, formation of the NICD-CSL-MAML complex and ubiquitination of Notch receptors are both key processes of Notch signaling,2,80–82 and blocking these aspects may not only affect Notch4, but also Notch1-3 signaling pathways. Thus, the efficiency and safety of these treatment methods should be further evaluated in future studies.
The Relationship Between Notch4 and Other Notch Receptors: An Uncertain Factor for Notch4-Targeting Treatment Therapies
Notch4 was found to act in conjunction with other Notch receptors in the cell, especially Notch1.83–85 Notch signal transduction assays performed by James et al revealed that Notch4 can inhibit Notch1 signaling in cultured NIH3T3 mouse embryonic fibroblasts and C2C12 mouse myoblasts.84 Mechanistically, the extracellular domain of Notch4 interacts with the unprocessed full-length Notch1 protein, which interferes with the subsequent S1 cleavage and trafficking of Notch1, thereby inhibiting Notch1 activation.84 In angiogenesis, activated Notch4 in ECs can positively regulate vascular maturation, while Notch1 is inactivated during this process.85 Notch1 decoys, novel soluble constructs based on the extracellular domain of Notch1, can block Notch4-induced EC morphogenesis and inhibit neo-angiogenesis.83 However, further details of the relationship between Notch1 and other Notch receptors are poorly investigated and still unknown.
In addition to Notch4, Notch1-3 receptors also play critical roles during cancer progression,6–8 and blocking Notch4 activation may cause potential effects on Notch1-3 receptors. Thus, further studies are needed to clarify the relationship between Notch4 and other Notch receptors before we can truly appreciate the potential risks and efficacy of Notch4-targeting therapies.
Discussion and Conclusion
As an important member of the Notch family, Notch4 has a unique role in regulating cellular behaviors in both tumor cells and tumor vasculature. Several studies have confirmed that Notch4 is a potential biomarker for CSCs. Accordingly, Notch4-positive tumor cells exhibit powerful stem cell-like self-renewal, invasion, and drug-resistance, which are mediated by complex mechanisms (See parts 3–5). In addition, Notch4 signaling activated by DLL4 has special regulatory effects on tumor vasculature, which includes inhibiting tumor neo-angiogenesis and promoting tumor vascular perfusion, as well as inducing tumor vessel remodeling and VM formation (See part 6). However, although the results of most of Notch4-related studies have shown that the activation of Notch4 signaling has oncogenic effects, tumor-suppressive mechanisms induced by Notch4 were also found.49,59 As suggested by Naik et al, the roles of Notch4 signaling in promoting or inhibiting tumor cell survival may depend on the cellular context,59 and the exact roles of Notch4 signaling in different types of cancer need further exploration.
Several potential strategies for inhibiting Notch4 expression or blocking Notch4 signal transduction in cancer have been applied in pre-clinical in vivo and in vitro experiments (See part 7). These include (a) knockdown of the Notch4 gene, (b) developing anti-Notch4 antibody drugs, (c) downregulation of Notch4 expression at the post-transcriptional level by ectopic expression of miRNAs, (d) targeting the γ-secretase complex by GSIs or Nicastrin inhibitors, (e) preventing the formation of the N4ICD-CSL-MAML complex in the nucleus, and (f) promoting the proteasomal degradation of Notch4 receptor or N4ICD. In addition, it is critical to investigate the relationship between Notch4 and other Notch receptors, as well as to estimate the potential effects of these Notch4-targeting strategies on other Notch receptors, both in vitro and in vivo.
In conclusion, aberrant Notch4 signaling contributes to cancer occurrence and progression by regulating diverse cellular behaviors, and several molecular mechanisms are involved in this process. Future studies are expected to further elucidate the various roles of Notch4 signaling in cancer, as well as the potential effects and clinical value of the application of different Notch4-targeting therapeutic strategies.
Funding Statement
There is no funding to report.
Abbreviations
CSL, CBF-1 (RBPJ)/suppressor of hairless/Lag1; MAML, Mastermind-like protein; FBXW7, F-box and WD repeat domain containing; DTX3, Deltex E3 ubiquitin ligase 3; OS, Over Survival; DFS, Disease Free Survival; ALDH1, Aldehyde dehydrogenase7; Oct4, Organic cation/carnitine transporter4; Sox2, SRY-box transcription factor 2; NF-κB, Nuclear factor kappa B; DCAF13, DDB1 and CUL4 associated factor 13; Stat3, Signal transducer and activator of transcription 3; MMP, Matrix metallopeptidase; PKCα, Protein kinase C alpha; NCL, Nucleolin; NPM, Nucleophosmin; FBL, Fibrillarin; ER, Endoplasmic Reticulum; TRAIL, TNF-related apoptosis-inducing ligand; Akt, Serine/threonine kinase 1; ROS, Reactive oxygen species; mTOR, mechanistic target of rapamycin kinase; ERK1/2, Extracellular regulated MAP kinase1/2; VEGF, Vascular Endothelial Growth Factor.
Data Sharing Statement
Not applicable.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Siebel C, Lendahl U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017 [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Sci STKE. 2006;2006:cm7. [DOI] [PubMed] [Google Scholar]
- 4.Carrieri FA, Dale JK. Turn It Down a Notch. Front Cell Develop Biol. 2016;4:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Li H, Mao A, et al. DCAF13 promotes triple-negative breast cancer metastasis by mediating DTX3 mRNA degradation. Cell Cycle. 2020;19(24):3622–3631. doi: 10.1080/15384101.2020.1859196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gharaibeh L, Elmadany N, Alwosaibai K, Alshaer W. Notch1 in Cancer Therapy: possible Clinical Implications and Challenges. Mol Pharmacol. 2020;98:559–576. doi: 10.1124/molpharm.120.000006 [DOI] [PubMed] [Google Scholar]
- 7.Xiu MX, Liu YM. The role of oncogenic Notch2 signaling in cancer: a novel therapeutic target. Am J Cancer Res. 2019;9:837–854. [PMC free article] [PubMed] [Google Scholar]
- 8.Aburjania Z, Jang S, Whitt J, Jaskula-Stzul R, Chen H, Rose JB. The Role of Notch3 in Cancer. oncologist. 2018;23:900–911. doi: 10.1634/theoncologist.2017-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiu MX, Liu YM, Kuang BH. The Role of DLLs in Cancer: a Novel Therapeutic Target. Onco Targets Ther. 2020;13:3881–3901. doi: 10.2147/OTT.S244860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiu MX, Liu YM, Kuang BH. The oncogenic role of Jagged1/Notch signaling in cancer. Biomed Pharmacother. 2020;129:110416. doi: 10.1016/j.biopha.2020.110416 [DOI] [PubMed] [Google Scholar]
- 11.Snijders AM, Schmidt BL, Fridlyand J, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601 [DOI] [PubMed] [Google Scholar]
- 12.Lunde MLS, Roman E, Warnakulasuriya S, et al. Profiling of chromosomal changes in potentially malignant and malignant oral mucosal lesions from South and South-East Asia using array-comparative genomic hybridization. Cancer Genomics Proteomics. 2014;11:127–140. [PubMed] [Google Scholar]
- 13.Hu Z-Y, Xie N, Tian C, et al. Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance. EBioMedicine. 2018;32:111–118. doi: 10.1016/j.ebiom.2018.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang X, Vacher S, Boulai A, et al. Targeted next-generation sequencing identifies clinically relevant somatic mutations in a large cohort of inflammatory breast cancer. Breast Cancer Res. 2018;20:88. doi: 10.1186/s13058-018-1007-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie M, Wei S, Wu X, Li X, You Y, He C. Alterations of Notch pathway in patients with adenoid cystic carcinoma of the trachea and its impact on survival. Lung Cancer (Amsterdam, Netherlands). 2018;121:41–47. doi: 10.1016/j.lungcan.2018.04.020 [DOI] [PubMed] [Google Scholar]
- 16.Cui M, Hu Y, Bi Y, et al. Preliminary exploration of potential molecular therapeutic targets in recurrent and metastatic parathyroid carcinomas. Int j Cancer. 2019;144:525–532. doi: 10.1002/ijc.31948 [DOI] [PubMed] [Google Scholar]
- 17.Ho AS, Ochoa A, Jayakumaran G, et al. Genetic hallmarks of recurrent/metastatic adenoid cystic carcinoma. J Clin Invest. 2019;129:4276–4289. doi: 10.1172/JCI128227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh J, Jeong CW, Choi S, et al. Sharing the initial experience of pan-cancer panel analysis in high-risk renal cell carcinoma in the Korean population. BMC Urol. 2020;20:125. doi: 10.1186/s12894-020-00687-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Duan N, Wang Y, et al. The clinicopathological and genetic features of ovarian diffuse large B-cell lymphoma. Pathology. 2020;52:206–212. doi: 10.1016/j.pathol.2019.09.014 [DOI] [PubMed] [Google Scholar]
- 20.Ahn S, Hyeon J, Park C-K. Notch1 and Notch4 are markers for poor prognosis of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2013;12:286–294. doi: 10.1016/S1499-3872(13)60046-6 [DOI] [PubMed] [Google Scholar]
- 21.Wu W-R, Shi X-D, Zhang R, et al. Clinicopathological significance of aberrant Notch receptors in intrahepatic cholangiocarcinoma. Int J Clin Exp Pathol. 2014;7(6):3272–3279. [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Sun B, Zhu D, et al. Notch4+ cancer stem-like cells promote the metastatic and invasive ability of melanoma. Cancer Sci. 2016;107:1079–1091. doi: 10.1111/cas.12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mk H, Prince S, Mohan AM, Krishnan KV, Devi A. Association of Notch4 with metastasis in human oral squamous cell carcinoma. Life Sci. 2016;156:38–46. doi: 10.1016/j.lfs.2016.05.019 [DOI] [PubMed] [Google Scholar]
- 24.Wang J-W, Wei X-L, Dou X-W, Huang W-H, Du C-W, Zhang G-J. The association between Notch4 expression, and clinicopathological characteristics and clinical outcomes in patients with breast cancer. Oncol Lett. 2018;15:8749–8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DC, Jin H, Lee JS, Son E, Lee GW, Kim HJ. P2Y(2)R has a significant correlation with Notch-4 in patients with breast cancer. Oncol Lett. 2020;20:647–654. doi: 10.3892/ol.2020.11630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L, Wang D, Sheng D, et al. NOTCH4 maintains quiescent mesenchymal-like breast cancer stem cells via transcriptionally activating SLUG and GAS1 in triple-negative breast cancer. Theranostics. 2020;10:2405–2421. doi: 10.7150/thno.38875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Liu W, Tang D, et al. Prognostic values of four Notch receptor mRNA expression in gastric cancer. Sci Rep. 2016;6:28044. doi: 10.1038/srep28044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu G, Chen Z, Li J, et al. NOTCH4 Is a Novel Prognostic Marker that Correlates with Colorectal Cancer Progression and Prognosis. J Cancer. 2018;9(13):2374–2379. doi: 10.7150/jca.26359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaik JP, Alanazi IO, Pathan AAK, et al. Frequent Activation of Notch Signaling Pathway in Colorectal Cancers and Its Implication in Patient Survival Outcome. J Oncol. 2020;2020:6768942. doi: 10.1155/2020/6768942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Yang R, Wang X, et al. Evaluation of the correlation of vasculogenic mimicry, Notch4, DLL4, and KAI1/CD82 in the prediction of metastasis and prognosis in non-small cell lung cancer. Medicine. 2018;97:e13817. doi: 10.1097/MD.0000000000013817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takam Kamga P, Dal Collo G, Resci F, et al. Notch Signaling Molecules as Prognostic Biomarkers for Acute Myeloid Leukemia. Cancers (Basel). 2019;11(12):1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasnauskienė A, Jonušienė V, Krikštaponienė A, et al. NOTCH1, NOTCH3, NOTCH4, and JAG2 protein levels in human endometrial cancer. Medicina. 2014;50:14–18. doi: 10.1016/j.medici.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 33.Violeta J, Ausra S, Nadezda L, et al. Down-regulated expression of Notch signaling molecules in human endometrial cancer. Med Onco. 2013;30:1–7. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Bu X, Yang J, et al. NOTCH4 regulates colorectal cancer proliferation, invasiveness, and determines clinical outcome of patients. J Cell Physiol. 2018;233:6975–6985. doi: 10.1002/jcp.26619 [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Wang X, Huang S, Wang L, Han L, Yu S. Prognostic roles of Notch receptor mRNA expression in human ovarian cancer. Oncotarget. 2017;8:32731–32740. doi: 10.18632/oncotarget.16387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison H, Farnie G, Howell SJ, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Angelo RC, Ouzounova M, Davis A, et al. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol Cancer Ther. 2015;14:779–787. doi: 10.1158/1535-7163.MCT-14-0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S-H, Singh SV. The role of polycomb group protein Bmi-1 and Notch4 in breast cancer stem cell inhibition by benzyl isothiocyanate. Breast Cancer Res Treat. 2015;149:681–692. doi: 10.1007/s10549-015-3279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apostolou P, Toloudi M, Ioannou E, et al. Study of the interaction among Notch pathway receptors, correlation with stemness, as well as their interaction with CD44, dipeptidyl peptidase-IV, hepatocyte growth factor receptor and the SETMAR transferase, in colon cancer stem cells. J Recept Signal Transduct Res. 2013;33:353–358. doi: 10.3109/10799893.2013.828072 [DOI] [PubMed] [Google Scholar]
- 40.Nguyen CH, Bauer K, Hackl H, et al. All-trans retinoic acid enhances, and a pan-RAR antagonist counteracts, the stem cell promoting activity of EVI1 in acute myeloid leukemia. Cell Death Dis. 2019;10:944. doi: 10.1038/s41419-019-2172-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rustighi A, Zannini A, Tiberi L, et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. 2014;6:99–119. doi: 10.1002/emmm.201302909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reipas KM, Law JH, Couto N, et al. Luteolin is a novel p90 ribosomal S6 kinase (RSK) inhibitor that suppresses Notch4 signaling by blocking the activation of Y-box binding protein-1 (YB-1). Oncotarget. 2013;4:329–345. doi: 10.18632/oncotarget.834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding LC, She L, Zheng DL, et al. Notch-4 contributes to the metastasis of salivary adenoid cystic carcinoma. Oncol Rep. 2010;24:363–368. [DOI] [PubMed] [Google Scholar]
- 44.Strizzi L, Margaryan NV, Gerami P, et al. Translational significance of Nodal, Cripto-1 and Notch4 in adult nevi. Oncol Lett. 2016;12:1349–1354. doi: 10.3892/ol.2016.4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Kuang Y, Yan W, Xu Q, Ren Q. Notch-4 silencing inhibits prostate cancer growth and EMT via the NF-κB pathway. Apoptosis. 2017;22:1–8. doi: 10.1007/s10495-017-1368-0 [DOI] [PubMed] [Google Scholar]
- 46.Fukusumi T, Guo TW, Sakai A, et al. The NOTCH4-HEY1 Pathway Induces Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res. 2018;24:619–633. doi: 10.1158/1078-0432.CCR-17-1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costa FF, Seftor EA, Bischof JM, et al. Epigenetically reprogramming metastatic tumor cells with an embryonic microenvironment. Epigenomics. 2009;1:387–398. doi: 10.2217/epi.09.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardy KM, Kirschmann DA, Seftor EA, et al. Regulation of the embryonic morphogen Nodal by Notch4 facilitates manifestation of the aggressive melanoma phenotype. Cancer Res. 2010;70:10340–10350. doi: 10.1158/0008-5472.CAN-10-0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonyadi RE, Hammerlindl H, Wels C, et al. Notch4 signaling induces a mesenchymal-epithelial-like transition in melanoma cells to suppress malignant behaviors. Cancer Res. 2016;76:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bui QT, Im JH, Jeong SB, et al. Essential role of Notch4/STAT3 signaling in epithelial-mesenchymal transition of tamoxifen-resistant human breast cancer. Cancer Lett. 2017;390:115–125. doi: 10.1016/j.canlet.2017.01.014 [DOI] [PubMed] [Google Scholar]
- 51.Fengyan Y, Yu J, Yinghua Z, et al. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem. 2012;287:465–473. doi: 10.1074/jbc.M111.280768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peiffer DS, Wyatt D, Zlobin A, et al. DAXX Suppresses Tumor-Initiating Cells in Estrogen Receptor-Positive Breast Cancer Following Endocrine Therapy. Cancer Res. 2019;79:4965–4977. doi: 10.1158/0008-5472.CAN-19-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gelsomino L, Panza S, Giordano C, et al. Mutations in the estrogen receptor alpha hormone binding domain promote stem cell phenotype through notch activation in breast cancer cell lines. Cancer Lett. 2018;428:12–20. doi: 10.1016/j.canlet.2018.04.023 [DOI] [PubMed] [Google Scholar]
- 54.Simões BM, O’Brien CS, Eyre R, et al. Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep. 2015;12:1968–1977. doi: 10.1016/j.celrep.2015.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lombardo Y, Filipović A, Molyneux G, et al. Nicastrin regulates breast cancer stem cell properties and tumor growth in vitro and in vivo. Proc Natl Acad Sci U S A. 2012;109:16558–16563. doi: 10.1073/pnas.1206268109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lombardo Y, Faronato M, Filipovic A, Vircillo V, Magnani L, Coombes RC. Nicastrin and Notch4 drive endocrine therapy resistance and epithelial to mesenchymal transition in MCF7 breast cancer cells. Breast Cancer Res. 2014;16(3):R62. doi: 10.1186/bcr3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun J, Pannuti A, Espinoza I, et al. Crosstalk between PKCα and Notch-4 in endocrine-resistant breast cancer cells. Oncogenesis. 2013;2:e60. doi: 10.1038/oncsis.2013.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saini N, Sarin A. Nucleolar localization of the Notch4 intracellular domain underpins its regulation of the cellular response to genotoxic stressors. Cell Death Discovery. 2020;6:7. doi: 10.1038/s41420-020-0242-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naik S, MacFarlane M, Sarin A. Notch4 Signaling Confers Susceptibility to TRAIL-Induced Apoptosis in Breast Cancer Cells. J Cell Biochem. 2015;116:1371–1380. doi: 10.1002/jcb.25094 [DOI] [PubMed] [Google Scholar]
- 60.Qian C-J, Chen -Y-Y, Zhang X, et al. Notch4 inhibition reduces migration and invasion and enhances sensitivity to docetaxel by inhibiting Akt/fascin in pancreatic cancer cells. Oncol Lett. 2016;12:3499–3505. doi: 10.3892/ol.2016.5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takam Kamga P, Dal Collo G, Midolo M, et al. Inhibition of Notch Signaling Enhances Chemosensitivity in B-cell Precursor Acute Lymphoblastic Leukemia. Cancer Res. 2019;79:639–649. doi: 10.1158/0008-5472.CAN-18-1617 [DOI] [PubMed] [Google Scholar]
- 62.Nwabo Kamdje AH, Bassi G, Pacelli L, et al. Role of stromal cell-mediated Notch signaling in CLL resistance to chemotherapy. Blood Cancer J. 2012;2:e73. doi: 10.1038/bcj.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Costa MJ, Wu X, Cuervo H, et al. Notch4 is required for tumor onset and perfusion. Vasc Cell. 2013;5(1):7. doi: 10.1186/2045-824X-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J-F, Chen Y, Qiu -X-X, et al. The vascular delta-like ligand-4 (DLL4)-Notch4 signaling correlates with angiogenesis in primary glioblastoma: an immunohistochemical study. Tumour Biol. 2016;37:3797–3805. doi: 10.1007/s13277-015-4202-8 [DOI] [PubMed] [Google Scholar]
- 65.Leong KG, Hu X, Li L, et al. Activated Notch4 inhibits angiogenesis: role of beta 1-integrin activation. Mol Cell Biol. 2002;22:2830–2841. doi: 10.1128/MCB.22.8.2830-2841.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacKenzie F, Duriez P, Larrivee B, et al. Notch4-induced inhibition of endothelial sprouting requires the ankyrin repeats and involves signaling through RBP-Jkappa. Blood. 2004;104:1760–1768. doi: 10.1182/blood-2003-12-4244 [DOI] [PubMed] [Google Scholar]
- 67.Williams CK, Li JL, Murga M, Harris AL, Tosato G. Up-regulation of the Notch ligand Delta-like 4 inhibits VEGF-induced endothelial cell function. Blood. 2006;107:931–939. doi: 10.1182/blood-2005-03-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hainaud P, Contrerès J-O, Villemain A, et al. The role of the vascular endothelial growth factor-Delta-like 4 ligand/Notch4-ephrin B2 cascade in tumor vessel remodeling and endothelial cell functions. Cancer Res. 2006;66:8501–8510. doi: 10.1158/0008-5472.CAN-05-4226 [DOI] [PubMed] [Google Scholar]
- 69.Zhang J, Qiao L, Liang N, et al. Vasculogenic mimicry and tumor metastasis. J BUON. 2016;21:533–541. [PubMed] [Google Scholar]
- 70.Cheng R, Cai XR, Ke K, Chen YL. Notch4 inhibition suppresses invasion and vasculogenic mimicry formation of hepatocellular carcinoma cells. J Huazhong Univ Sci Technolog Med Sci. 2017;37:719–725. [DOI] [PubMed] [Google Scholar]
- 71.Bao S, Jin S, Wang C, Tu P, Hu K, Lu J. Androgen receptor suppresses vasculogenic mimicry in hepatocellular carcinoma via circRNA7/miRNA7-5p/VE-cadherin/Notch4 signalling. J Cell Mol Med. 2020. doi: 10.1111/jcmm.16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagamatsu I, Onishi H, Matsushita S, et al. NOTCH4 is a potential therapeutic target for triple-negative breast cancer. Anticancer Res. 2014;34:69–80. [PubMed] [Google Scholar]
- 73.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 74.Long LM, Zhan JK, Wang HQ, Li S, Chen YY, Liu YS. The Clinical Significance of miR-34a in Pancreatic Ductal Carcinoma and Associated Molecular and Cellular Mechanisms. Pathobiology. 2017;84:38–48. doi: 10.1159/000447302 [DOI] [PubMed] [Google Scholar]
- 75.Cheung CM, Lun WM, Chung TY, et al. MicroRNA-183 suppresses cancer stem-like cell properties in EBV-associated nasopharyngeal carcinoma. Bmc Cancer. 2016;16:495. doi: 10.1186/s12885-016-2525-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yutaka H, Yoshimitsu A, Takeshi O, Shu S, Yasuhito Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 2010;31:777–784. doi: 10.1093/carcin/bgq013 [DOI] [PubMed] [Google Scholar]
- 77.Li WJ, Xie XX, Bai J, Wang C, Zhao L, Jiang DQ. Increased expression of miR-1179 inhibits breast cancer cell metastasis by modulating Notch signaling pathway and correlates with favorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22:8374–8382. [DOI] [PubMed] [Google Scholar]
- 78.Nwabo Kamdje AH, Mosna F, Bifari F, et al. Notch-3 and Notch-4 signaling rescue from apoptosis human B-ALL cells in contact with human bone marrow-derived mesenchymal stromal cells. Blood. 2011;118:380–389. doi: 10.1182/blood-2010-12-326694 [DOI] [PubMed] [Google Scholar]
- 79.López-Nieva P, González-Sánchez L, Cobos-Fernández M, Córdoba R, Santos J, Fernández-Piqueras J. More Insights on the Use of γ-Secretase Inhibitors in Cancer Treatment. oncologist. 2021;26:e298–e305. doi: 10.1002/onco.13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okubo Y, Sugawara T, Abe-Koduka N, Kanno J, Kimura A, Saga Y. Lfng regulates the synchronized oscillation of the mouse segmentation clock via trans-repression of Notch signalling. Nat Commun. 2012;3:1141. doi: 10.1038/ncomms2133 [DOI] [PubMed] [Google Scholar]
- 81.Kar R, Jha SK, Ojha S, et al. The FBXW7-NOTCH interactome: a ubiquitin proteasomal system-induced crosstalk modulating oncogenic transformation in human tissues. Cancer Rep (Hoboken, NJ). 2021;e1369{EPub]. doi: 10.1002/cnr2.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dutta D, Sharma V, Mutsuddi M, Mukherjee A. Regulation of Notch signaling by E3 ubiquitin ligases. FEBS J. 2021. doi: 10.1111/febs.15792 [DOI] [PubMed] [Google Scholar]
- 83.Funahashi Y, Hernandez SL, Das I, et al. A notch1 ectodomain construct inhibits endothelial notch signaling, tumor growth, and angiogenesis. Cancer Res. 2008;68:4727–4735. doi: 10.1158/0008-5472.CAN-07-6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.James AC, Szot JO, Iyer K, et al. Notch4 reveals a novel mechanism regulating Notch signal transduction. Biochim Biophys Acta. 2014;1843:1272–1284. doi: 10.1016/j.bbamcr.2014.03.015 [DOI] [PubMed] [Google Scholar]
- 85.Pedrosa AR, Trindade A, Fernandes AC, et al. Endothelial Jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler Thromb Vasc Biol. 2015;35:1134–1146. doi: 10.1161/ATVBAHA.114.304741 [DOI] [PubMed] [Google Scholar]
- 86.Donnem T, Andersen S, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer. 2010;116:5676–5685. doi: 10.1002/cncr.25551 [DOI] [PubMed] [Google Scholar]