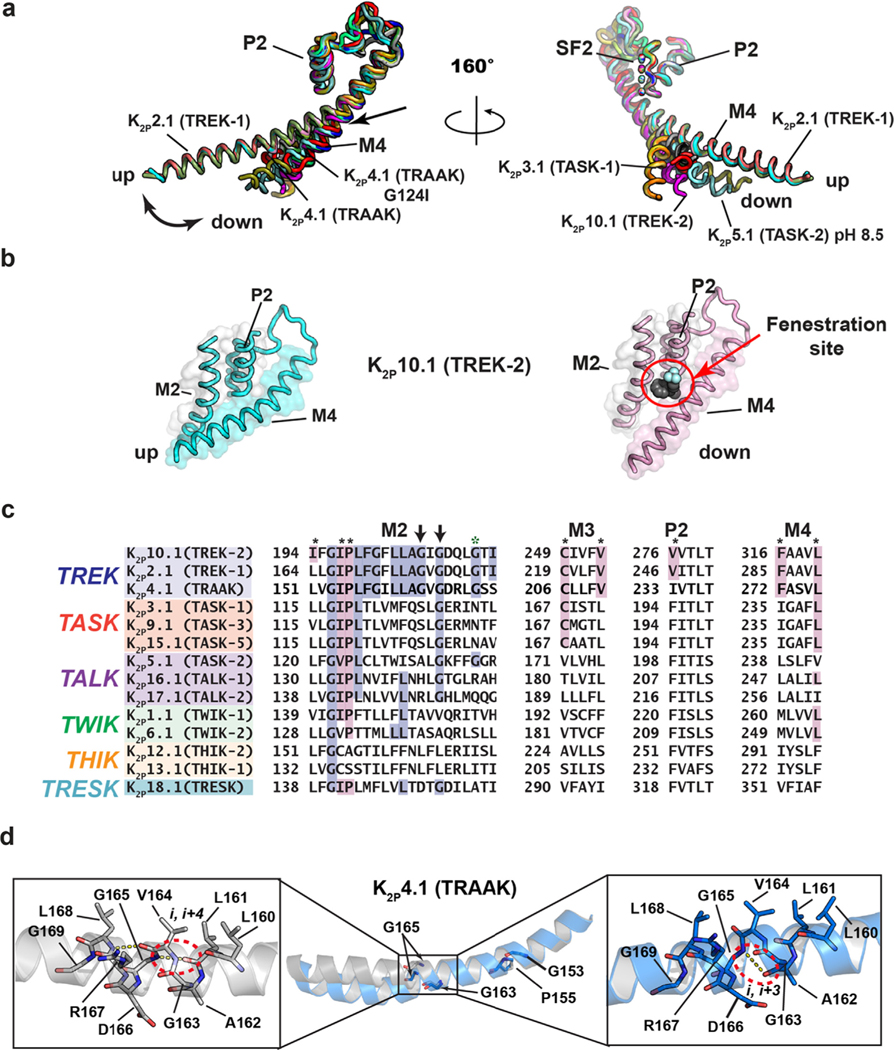
Figure 4.
K2P transmembrane moving parts. (a) Superposition of the P2-SF2-M4 portion for: K2P1.1 (TWIK-1) (3UKM)36 (red), K2P2.1 (TREK-1) (6CQ6)37 (smudge), K2P2.1 (TREK-1):ML335 (6CQ8)37 (deep salmon), K2P2.1 (TREK-1):ML402 (6CQ9),37 K2P3.1 (TASK-1) (6RV2) (orange),38 K2P3.1 (TASK-1):BAY1000493 (6RV3) (yellow orange),38 K2P3.1 (TASK-1):BAY2341237(6RV4) (olive),38 K2P4.1 (TRAAK) (3UM7)39 (aquamarine), (4I9W)56 (limon), (4WFE) (forest green),58 (4WFF) (white),58 (4WFG) (grey),58 (4WFH) (black),58 K2P4.1 (TRAAK) G124I (4RUE) (blue),57 K2P4.1 (TRAAK) W262S (4RUF) (lime green),57 K2P5.1 (TASK-2) pH 6.5 (6WLV) (deep olive),41 K2P5.1 (TASK-2) pH 8.5 (6WM0) (light teal),41 K2P10.1 (TREK-2) (4BW5)40 (pink), (4XDJ)40 (magenta), (4XDK)40 (purple). Select structures are indicated. Arrow indicates point of M4 bend. (b) View of the ‘up’ (left) and ‘down’ (right) M4 conformations in K2P10.1 (TREK-2). Fenestration site is occupied by norfluoxetine (space filling) and is indicated by the red circle. (c) Sequence comparisons of the M2 helix and elements that frame the Fenestration site. Residues that interact with the Fenestration site ligand are indicated by the black asterisks and are highlighted in magenta (cf. Figure 6). TREK subfamily ‘GVG’ sequence is indicated by the arrows. Green asterisk indicates the position of azo-isoflurane labeling.143 (d) Buckling at the conserved TREK family ‘GVG’ sequence in K2P4.1 (TRAAK). Dashed red oval highlights the site of the i,i + 4 to i,i + 3 hydrogen bond shift. Residue numbers are from human K2P4.1 (TRAAK) PDB:4RUE.57 Genbank sequences in ‘c’ are the same as those in Figure 2(a).