Dear Editor,
A number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfections [1–5] have been communicated since the first reported case in August 2020 [6]. Confirmation of reinfection has always been based on differences between sequential SARS-CoV-2 strains in the first and second episodes, either major, different lineages/a large number of single nucleotide polymorphisms (SNPs) or moderate, lower diversity, but still relevant, between the strains.
A recurrence is not classified as reinfection without the support of viral genomic data; however, there is a lack of genomic rigor when documenting whether the samples used to characterize the strains involved in the sequential episodes correspond to the same patient. This is essential to support viral genomics findings and demonstrate that different SARS-CoV-2 strains have sequentially infected a single patient. To the best of our knowledge, only 2 SARS-CoV-2 reinfection studies have taken this point into consideration; these have shown, supported by short tandem repeat (STR) analysis, identical host markers for the nasopharyngeal samples collected for the analyses [4, 7].
With the striking increase in laboratory workload once the first coronavirus disease 2019 (COVID-19) wave hit, collecting sample remnants for future studies was a real challenge. Mistakes in sample labeling or in aliquoting were more likely to occur under the experienced stress. Thus, when documenting a reinfection in COVID-19, genomic data ensuring that the stored sample belongs to the same patient should be a requirement before a SARS-CoV-2 reinfection report is admitted as such.
Here, we present a COVID-19 recurrence case that met all the clinical and microbiological requirements of a SARS-CoV-2 reinfection. The patient is a 61-year-old man with a history of chronic obstructive pulmonary disease, obesity, and ankylosing spondylitis, who had a first COVID-19 episode in March 2020 (positive SARS-CoV-2 reverse transcription polymerase chain reaction [RT-PCR] on March 17; cycle threshold [Ct] 19) and a second episode 6 months later, in September (positive RT-PCRs on September 13, 17, 28, and October 4; Ct values 17, 18, 23, and 35, respectively). Both episodes were characterized by mild symptoms; the second occurred in the context of a SARS-CoV-2 nosocomial infection. Three negative SARS-CoV-2 RT-PCRs were obtained between the first and second episodes (March 30, August 4, and September 3). Serology was not available for the first episode, but it was negative in August, and 10 days after the second episode, the patient showed a positive antibody titer.
Genomic viral analyses determined 6 SNPs for the SARS-CoV-2 strain, lineage 19A, obtained from the first episode, with respect to the Wuhan-1 reference sequence (Figure 1A). The strain from the second episode did not show any of the SNPs for the first episode strain and carried 16 other SNPs (Figure 1A, Table 1) and belonged to a different lineage, namely 20A.EU1, the primary lineage in Spain and many European countries since the end of June [8]. Based on major genomic differences between the 2 strains, we labeled the recurrence a reinfection.
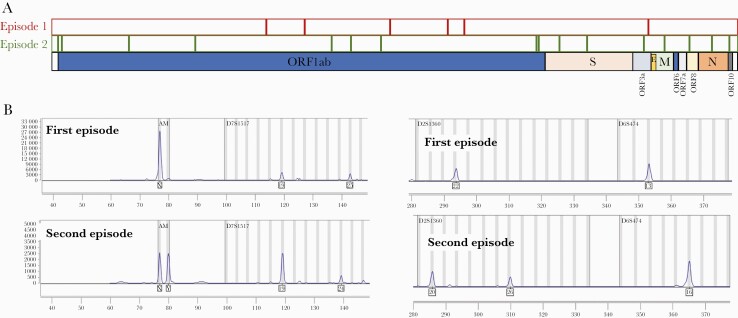
Figure 1.
A, Schematic single nucleotide polymorphism distribution (vertical black lines) along the SARS-CoV-2 chromosome for the 2 COVID-19 episodes. B, Human identity testing analysis was done by short tandem repeat PCR (Mentype Chimera Biotype, Germany) on the same samples used to perform the SARS-CoV-2 RT-PCR, and they were subsequently sequenced. Representative loci of the 12 noncoding short tandem repeat loci and the gender-specific locus amelogenin were analyzed. They were labeled with 3 different dyes (6-FAM, BTG, or BTY) using the Mentype Chimera PCR amplification kit (Biotype, Germany). Abbreviations: COVID-19, coronavirus disease 2019; PCR, polymerase chain reaction; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 1.
Differential SNPs Between Episodes
| 03/17/2020 | 09/13/2020 | ||||
|---|---|---|---|---|---|
| Position | First Episode | Second Episode | |||
| C|9360|T | 1 | 0 | ORF1ab | missense_variant | Thr3032Ile |
| G|11083|T | 1 | 0 | ORF1ab | missense_variant | Leu3606Phe |
| C|14805|T | 1 | 0 | ORF1ab | synonymous_variant | Tyr4847Tyr |
| T|17247|C | 1 | 0 | ORF1ab | synonymous_variant | Arg5661Arg |
| A|18077|G | 1 | 0 | ORF1ab | missense_variant | Lys5938Arg |
| G|26144|T | 1 | 0 | ORF3a | missense_variant | Gly251Val |
| C|241|T | 0 | 1 | ORF1ab | upstream_gene_variant | -25C > T |
| T|445|C | 0 | 1 | ORF1ab | synonymous_variant | Val60Val |
| C|3037|T | 0 | 1 | ORF1ab | synonymous_variant | Phe924Phe |
| C|6286|T | 0 | 1 | ORF1ab | synonymous_variant | Thr2007Thr |
| C|12119|T | 0 | 1 | ORF1ab | missense_variant | Pro3952Ser |
| C|13115|T | 0 | 1 | ORF1ab | synonymous_variant | Leu4284Leu |
| C|14408|T | 0 | 1 | ORF1ab | missense_variant | Pro4715Leu |
| A|21222|T | 0 | 1 | ORF1ab | synonymous_variant | Ala6986Ala |
| G|21255|C | 0 | 1 | ORF1ab | synonymous_variant | Ala6997Ala |
| C|22227|T | 0 | 1 | S | missense_variant | Ala222Val |
| A|23403|G | 0 | 1 | S | missense_variant | Asp614Gly |
| C|25889|T | 0 | 1 | ORF3a | missense_variant | Ser166Leu |
| C|26801|G | 0 | 1 | M | synonymous_variant | Leu93Leu |
| C|27944|T | 0 | 1 | ORF8 | synonymous_variant | His17His |
| C|28932|T | 0 | 1 | N | missense_variant | Ala220Val |
| G|29645|T | 0 | 1 | ORF10 | missense_variant | Val30Leu |
Abbreviation: SNP, single nucleotide polymorphism.
We then performed STR analysis from the same total nucleic acid preparation that had been used to perform the viral genomic analysis (Supplementary Data). It revealed a different origin of the specimens due to completely different patterns for the first- and second-episode specimens (Figure 1B). In fact, the material from the first episode belonged to a woman and the second to a man, indicating mislabeling or mishandling of the specimen assigned to our (male) patient’s first episode. The certainty of our patient’s second episode was proved by (i) 4 sequential positive RT-PCRs, (ii) 0 SNPs between the sequences from 2 of the SARS-CoV-2-positive specimens from his second episode, and (iii) STR analysis indicating that these last 2 specimens belonged to the same person (both males). The STR analysis revealed that the reinfection was wrongly classified as reinfection, something that would have gone unnoticed if the host material had not been characterized.
Our data highlight the potential incorrect classification of SARS-CoV-2 infections when samples from COVID-19 sequential episodes, used for viral genomic sequencing, are not confirmed to belong to the same individual by host genetic analysis. STR-based analysis should be mandatory in all reports assessing SARS-CoV-2 reinfections.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We are grateful to Dainora Jaloveckas (cienciatraducida.com) for editing and proofreading assistance.
Financial support. This work was supported by Instituto de Salud Carlos III (Ref COV20/00140: SeqCOVID—Consorcio para la epidemiología genómica de SARS-CoV-2 en España) and by Consejo Superior de Investigaciones Científicas (CSIC; PTI Salud Global). L.P.L. has a Miguel Servet Contract (CPII20/00001).
Potential conflicts of interest. The authors declare no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. Laura Pérez Lago, Marina Machado: data analysis, MS revision. Marta Herranz: investigation. Pedro J. Sola-Campoy: bioinformatics. Julia Suárez-González: viral sequencing. Carolina Martínez-Laperche: host genetic analysis. Iñaki Comas, Patricia Muñoz: resources, MS revision. Luis Alcalá, Pilar Catalán: data analysis, databases, MS revision. Darío García de Viedma: conceptualization, data analysis, MS writing.
Patient consent. The patient’s written consent was obtained. The design of the work was approved by the Ethics Committee from Gregorio Marañón Hospital (reference: MICRO.HGUGM0.2020–042).
Data summary. BAM files from the sequences were deposited in ENA (EMBL; with human sequences already filtered out) under reference PRJEB43221.
Gregorio Marañón Microbiology-ID COVID 19 Study Group. Adán-Jiménez (Javier), Alcalá (Luis), Aldámiz (Teresa), Alonso (Roberto), Álvarez (Beatriz), Álvarez-Uría (Ana), Arias (Alexi), Arroyo (Luis Antonio), Berenguer (Juan), Bermúdez (Elena), Bouza (Emilio), Burillo (Almudena), Candela (Ana), Carrillo (Raquel), Catalán (Pilar), Cercenado (Emilia), Cobos (Alejandro), Díez (Cristina), Escribano (Pilar), Estévez (Agustín), Fanciulli (Chiara), Galar (Alicia), García (Mª Dolores), García de Viedma (Darío), Gijón (Paloma), González (Adolfo), Guillén (Helmuth) Guinea (Jesús), Haces (Laura Vanessa), Herranz (Marta), Kestler (Martha), López (Juan Carlos), Losada (Carmen Narcisa), Machado (Marina), Marín (Mercedes), Martín (Pablo), Montilla (Pedro), Moure (Zaira), Muñoz (Patricia), Olmedo (María), Padilla (Belén), Palomo (María), Parras (Francisco), Pérez-Granda (María Jesús), Pérez-Lago (Laura), Pérez (Leire), Pescador (Paula), R. Maus (Sandra), Reigadas (Elena), Rincón (Cristina), Rodríguez (Belén), Rodríguez (Sara), Rodríguez-Grande (Cristina), Rojas (Adriana), Ruiz-Serrano (María Jesús), Sánchez (Carlos), Sánchez (Mar), Serrano (Julia), Sola Campoy (Pedro J), Tejerina (Francisco), Valerio (Maricela), Veintimilla (Mª Cristina), Vesperinas (Lara), Vicente (Teresa), de la Villa (Sofía).
Contributor Information
Gregorio Marañón Microbiology-ID COVID-19 Study Group:
Javier Adán-Jiménez, Luis Alcalá, Teresa Aldámiz, Roberto Alonso, Beatriz Álvarez, Ana Álvarez-Uría, Alexi Arias, Luis Antonio Arroyo, Juan Berenguer, Elena Bermúdez, Emilio Bouza, Almudena Burillo, Ana Candela, Raquel Carrillo, Pilar Catalán, Emilia Cercenado, Alejandro Cobos, Cristina Díez, Pilar Escribano, Agustín Estévez, Chiara Fanciulli, Alicia Galar, Mª Dolores García, Darío García de Viedma, Paloma Gijón, Adolfo González, Helmuth Guillén, Jesús Guinea, Laura Vanessa Haces, Marta Herranz, Martha Kestler, Juan Carlos López, Carmen Narcisa Losada, Marina Machado, Mercedes Marín, Pablo Martín, Pedro Montilla, Zaira Moure, Patricia Muñoz, María Olmedo, Belén Padilla, María Palomo, Francisco Parras, María Jesús Pérez-Granda, Laura Pérez-Lago, Leire Pérez, Paula Pescador, Sandra R Maus, Elena Reigadas, Cristina Rincón, Belén Rodríguez, Sara Rodríguez, Cristina Rodríguez-Grande, Adriana Rojas, María Jesús Ruiz-Serrano, Carlos Sánchez, Mar Sánchez, Julia Serrano, Pedro J Sola Campoy, Francisco Tejerina, Maricela Valerio, Mª Cristina Veintimilla, Lara Vesperinas, Teresa Vicente, and Sofía de la Villa
References
- 1.Gupta V, Bhoyar RC, Jain A, et al. . Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clinical Infect Dis 2020. doi: 10.1093/cid/ciaa1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larson D, Brodniak SL, Voegtly LJ, et al. . A case of early re-infection with SARS-CoV-2. Clinical Infect Dis 2020. doi: 10.1093/cid/ciaa1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulder M, van der Vegt D, Oude Munnink BB, et al. . Reinfection of SARS-CoV-2 in an immunocompromised patient: a case report. Clinical Infect Dis 2020. doi: 10.1093/cid/ciaa1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richard L, Tillett JRS, Hartley PD, et al. . Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021 . Jan;21(1):52-58. doi: 10.1016/S1473-3099(20)30764-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Elslande J, Vermeersch P, Vandervoort K, et al. . Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis 2021; . 73:354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To KK, Hung IF, Ip JD, et al. . COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Kim S, Kim T, et al. . Evidence of severe acute respiratory syndrome coronavirus 2 reinfection after recovery from mild coronavirus disease 2019. Clinical Infect Dis 2020. doi: 10.1093/cid/ciaa1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodcroft E, Zuber M, Nadeau S, et al. . Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 2021; 595:707–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.