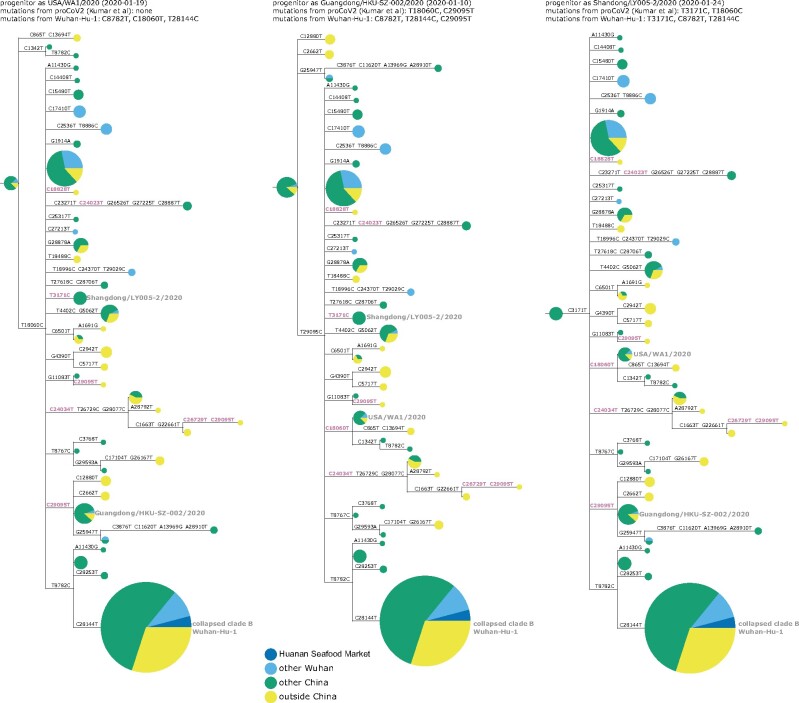
Fig. 3.
Phylogenetic trees of SARS-CoV-2 sequences in GISAID collected before February 2020. The trees are identical except they are rooted to make the progenitor each of the three sequences with highest identity to the RaTG13 bat coronavirus outgroup. Nodes are shown as pie charts with areas proportional to the number of observations of that sequence and colored by where the viruses were collected. The mutations on each branch are labeled, with mutations toward the nucleotide identity in the outgroup in purple. The labels at the top of each tree give the first known virus identical to each putative progenitor, as well as mutations in that progenitor relative to proCoV2 (Kumar et al. 2021) and Wuhan-Hu-1. The monophyletic group containing C28144T is collapsed into a node labeled “clade B” in concordance with the naming scheme of Rambaut et al. (2020); this clade contains Wuhan-Hu-1. Singleton mutations (mutations observed only once in the sequence set) are removed as described in more detail in the Methods. Supplementary figs. S4 and S5, Supplementary Material online, show identical results are obtained if the outgroup is RpYN06 or RmYN02.