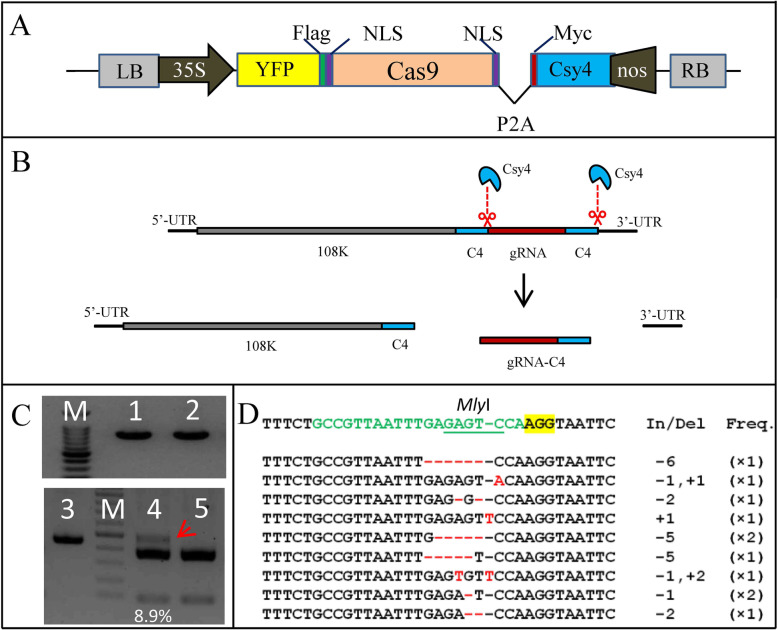
Fig. 3.
Cas9-induced genome editing with CCAC. (A) Schematic of Cas9-Cys4 construct. The YFP protein was fused to Cas9, while the Csy4 protein was separated by 2 A self-cleaving peptide (P2A). (B) gRNA release by Csy4 processing. The gRNA flanked by C4 sites, was replicated and moved along with ALSV virus RNAs. Csy4 cleaves and releases gRNA from the virus RNAs. The C4 site remains at the 3’ end of the gRNA. (C) Genome editing of PDS with CCAC. The photobeached leaves of “CCAC:vPDS+gPDS” plants in Figure 2B were or were not (as control) infiltrated with Cas9-Cys4 construct. 2dpi the genomic DNA was extracted and used as PCR template with primers flanking PDS locus. Lane 1 and lane 3 were the PCR products with photobleached leaves infiltrated by Cas9-Cys4 construct as template, while in lane 2 the template was the photobleached leaves without Cas9-Cys4 construct infiltration as control. Lanes4 and 5 show the MlyI -digested PCR products from lanes 1 and 2, respectively. The mutation rate (8.9%) was calculated by ImageJ. The red arrow indicates the MlyI -resistant band. M = DNA marker. Uncropped full-length gels of Figure 3C is shown in Supplementary Figure 9. (D) Alignment of PDS sequences with Cas9-induced indels obtained from the red arrow indicated band in C. The wild type sequence is shown at the top. The sequence targeted by gPDS is shown in green, whereas the mutations varying from that sequence are shown in red. PAM is highlighted in yellow. The total length (as indicated by number of bases) of indels (In/Del) and the frequency with which each DNA sequence pattern was observed (Freq.) are presented to the right of the sequences