Abstract
Background
Colorectal cancer (CRC) is a malignancy of the large intestine, whose development and prognosis have been demonstrated to be associated with altered lipid metabolism. High cholesterol intake is associated with an increased risk of CRC, and elevated serum cholesterol levels are known to be correlated with risk of developing CRC. Niemann-Pick C1-Like 1 (NPC1L1), a target of ezetimibe, plays an essential role in the absorption of intestinal cholesterol. However, whether the altered expression of NPC1L1 affects CRC development and prognosis is currently unknown.
Methods
Data corresponding to patients with CRC were obtained from The Cancer Genome Atlas (TCAG). Datasets from the Genome Data Analysis Center (GDAC) platform were analyzed to compare the expression of NPC1L1 in normal and CRC tissues using the Mann–Whitney U test and chi-square test. Further, the datasets from the Gene Expression Omnibus (GEO) database were analyzed. The log-rank test and multivariate Cox proportional hazard regression analysis were performed to determine whether NPC1L1 significantly affects the prognosis of CRC.
Results
The expression of NPC1L1 was found to be upregulated in CRC and was significantly associated with the N and pathological stages but not with the histological type, age, and sex. Increased NPC1L1 expression in CRC was related to poor patient survival, as evidenced by the Kaplan–Meier and multivariate regression analyses.
Conclusions
As high expression of NPC1L1 was associated with CRC development, pathological stage, and prognosis, NPC1L1 can serve as an independent prognostic marker for CRC.
Keywords: Niemann-Pick C1-Like 1, Ezetimibe, Cholesterol, Colorectal cancer, Prognosis, Overall survival, Stage, Molecular marker
Background
Colorectal cancer (CRC), a malignancy of the colon and rectum, is the third most common cancer and the fourth leading cause of cancer-related deaths worldwide [1]. The incidence of CRC is rapidly increasing in countries with medium-to-high human development indices, including those in Eastern Europe, South America, and Asia, owing to changes in dietary habits and westernized lifestyles [2, 3]. The incidence of CRC is 19.7 per 100,000 people, and the 5-year survival rate of patients with stage IV or metastatic CRC is 12 % [4]. Age, family history, inflammatory bowel disease (IBD), hereditary CRC, obesity, and diabetes are known risk factors for CRC. Despite the advances in our understanding of CRC pathogenesis, the response of advanced CRC to conventional chemotherapy is poor. Therefore, it is imperative to identify new molecular markers for predicting the prognosis of CRC, thereby improving the survival rates of the afflicted patients.
Many studies have reported that the development and progression of cancers are associated with altered cholesterol metabolism [5]. Oxysterol 27-hydroxycholesterol, a primary metabolite of cholesterol, was found to promote breast cancer progression in mouse models [6], and the level of total cholesterol was positively associated with the incidence of prostate cancer in men [7]. High levels of cholesterol intake are associated with an increased risk of CRC [8], and elevated serum cholesterol levels are correlated with CRC risk [9]. Among the molecules involved in cholesterol metabolism, Niemann-Pick C1-Like 1 (NPC1L1) is a transmembrane protein that is essential for the intestinal absorption of cholesterol [10]. Importantly, NPC1L1 is a target of ezetimibe, a drug that is used to treat dyslipidemia that cannot be managed using statin treatment. NPC1L1 is expressed at high levels in the human small intestine and liver but is expressed at low levels in the colon, kidneys, and brain [11]. NPC1L1 knockdown was reported to result in significantly decreased the number of tumors in a murine model of colitis-associated CRC [12]. However, whether the altered expression of NPC1L1 correlates with the development and prognosis of CRC in humans remains unclear.
In the present study, NPC1L1 expression in normal and CRC tissues was compared and a correlation was investigated between the expression of NPC1L1 and clinical characteristics. Moreover, the influence of NPC1L1 on the overall survival (OS) of CRC patients was examined, and the value of NPC1L1 as an independent prognostic factor for CRC was determined.
Methods
Patients and gene expression data
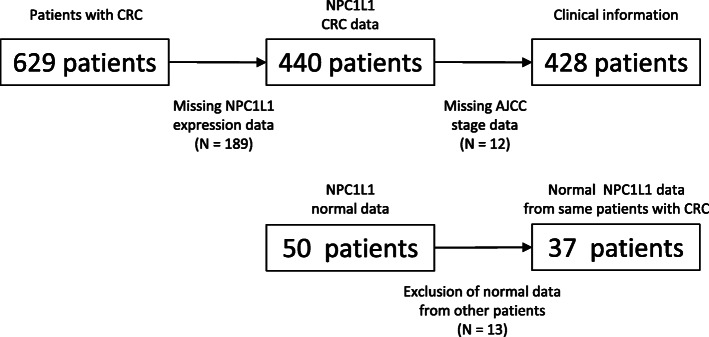
The Cancer Genome Atlas (TCGA) hosts large clinical datasets with information on DNA methylation, microRNAs and RNA expression in various types of cancers, including CRC. TCGA data corresponding to 629 patients with CRC were obtained as previously described [13]. Among these, data corresponding to 201 patients were excluded from the study because information regarding NPC1L1 expression for 189 patients and pathological stages for 12 patients was not available. Thus, data corresponding to a total of 428 patients with CRC were finally included for prognosis analysis in this study (Fig. 1). Histological type, age, race, TNM stage, and pathological stage were selected as the clinical variables. However, race as a variable was not eventually used for analysis because no data were available for 36.7 % of the patients whose data were curated in these datasets (Table 1). TNM staging is a system for classifying malignancies, wherein the T stage indicates the size of the primary tumor and the degree of invasion, the N stage indicates the extent to which the tumor has spread to the surrounding lymph nodes, and the M stage indicates the degree of metastasis to other organs. mRNA expression is the read-out of the total number of overlapping reads encompassing a gene during differential expression analysis in RNA sequencing, also known as the read count.
Fig. 1.
Flow chart for obtaining information for CRC patients with normal NPC1L1 expression
Table 1.
Clinical characteristics of patients with CRC
| Patient characteristics (N = 428) | Total (%) | |
|---|---|---|
| Histological type | Colon adenocarcinoma | 363 (84.8) |
| Colon mucinous adenocarcinoma | 60 (14) | |
| Unknown | 5 (1.2) | |
| Overall survival months (mean ± SDa) | 29.43 ± 26.08 | |
| Age (mean ± SDa) | 66.63 ± 13.08 | |
| Sex | Male | 228 (53.3) |
| Female | 200 (46.7) | |
| Race | American Indian or Alaska native | 1 (0.2) |
| Asian | 11 (2.6) | |
| Black or American | 57 (13.3) | |
| White | 202 (47.2) | |
| Unknown | 157 (36.7) | |
| T stage | Tis | 1 (0.2) |
| T1-T2 | 81 (18.9) | |
| T3-T4 | 346 (80.8) | |
| M stage | M0 | 321 (75) |
| M1 | 61 (14.3) | |
| Unknown | 46 (10.7) | |
| N stage | N0 | 250 (58.4) |
| N1-N2 | 178 (41.6) | |
| AJCC stage | Stage I | 72 (16.8) |
| Stage II | 169 (39.5) | |
| Stage III | 126 (29.4) | |
| Stage IV | 61 (14.3) | |
aSD standard deviation, CRC colorectal cancer, AJCC American Joint Committee on Cancer
Overall, 50 normal datasets comprising data corresponding to NPC1L1 expression were obtained from the Genome Data Analysis Center (GDAC) platform. Of the 50 normal datasets, 37 datasets containing information regarding NPC1L1 expression in CRC patients were included in this study. Owing to the fact that the definition of “normal” in the context of expression is based on the level of mRNA expression in the tissues adjacent to the CRC tissues, the expression of NPC1L1 in normal and CRC tissues from the same patients could be compared. When comparing the relative expression of NPC1L1 in paired tissues, expression values of NPC1L1 in each of the 35 tumor tissues were divided by those in paired normal tissues and expressed as the Log2 of expression. Two normal datasets were not used because the expression of NPC1L1 in two tumor tissues (calculated as numerator) was 0.
To confirm the expression and prognostic significance of NPC1L1 in CRC, NPC1L1 expression data from the Gene Expression Omnibus (GEO) datasets (GSE9348, GSE17536, and GSE129451) were obtained and analyzed.
Statistical analysis
The differences between the expression of NPC1L1 in normal and CRC tissues were examined using the Mann–Whitney U test after subjecting the data to the Shapiro–Wilk normality test as the data were not normally distributed. Statistical significance was set at P < 0.05. A chi-square test was performed to analyze the correlation between NPC1L1 expression and clinical characteristics. Overall survival (OS) was calculated from the date of diagnosis to death or last follow-up. For the Kaplan–Meier (KM) curves, patients with CRC were assigned to either the NPC1L1 low (n = 214) or NPC1L1 high (n = 214) expression group based on the median levels of NPC1L1 expression. The log-rank test was used to calculate the P values. Univariate and multivariate Cox proportional hazard regression analyses were performed to determine whether NPC1L1 is a significant predictor of OS in patients with CRC. Box and whisker plot construction, KM curve construction, and statistical analysis were performed using the Excel (2016) and SPSS programs (version 20).
Results
Baseline characteristics
The characteristics of the patients with CRC are summarized in Table 1. The mean age and OS of patients with CRC were 66.63 ± 13.08 years and 29.43 ± 26.08 months, respectively. Most of the CRC tissues were found to be adenocarcinoma (n = 363, 84.8 %), and the ratio between the number of males and females was not notably different (male: n = 228, female: n = 200). The proportion of Caucasian subjects with CRC was relatively high (n = 202, 47.2 %), and the proportion of Asian subjects with CRC was 2.6 %. Based on the American Joint Committee on Cancer (AJCC) staging system, the proportion of patients with stage I, II, III, and IV CRC was found to be 16.8 %, 39.5 %, 29.4 %, and 14.3 %, respectively.
Expression of NPC1L1 in normal and CRC tissues
NPC1L1 expression was found to be significantly higher in CRC tissues than that in normal tissues (normal: mean 7.00, CRC: mean 130.09, P < 0.05) (Fig. 2a). For accurate verification, the relative expression of NPC1L1 was analyzed in paired normal and tumor tissues from patients with CRC (Fig. 2b). NPC1L1 expression was higher in most CRC tissues than that in paired normal tissues. To determine whether the NPC1L1 expression levels in data—from databases other than TCGA—corresponding to normal and tumor tissues were consistent with the results obtained using TCGA data, a GEO dataset (GSE9348) was analyzed (Fig. 2c). The results indicated that the expression of NPC1L1 was significantly higher in CRC tissues than that in normal tissues (normal: mean 22.69, CRC: mean 81.35, P < 0.05).
Fig. 2.
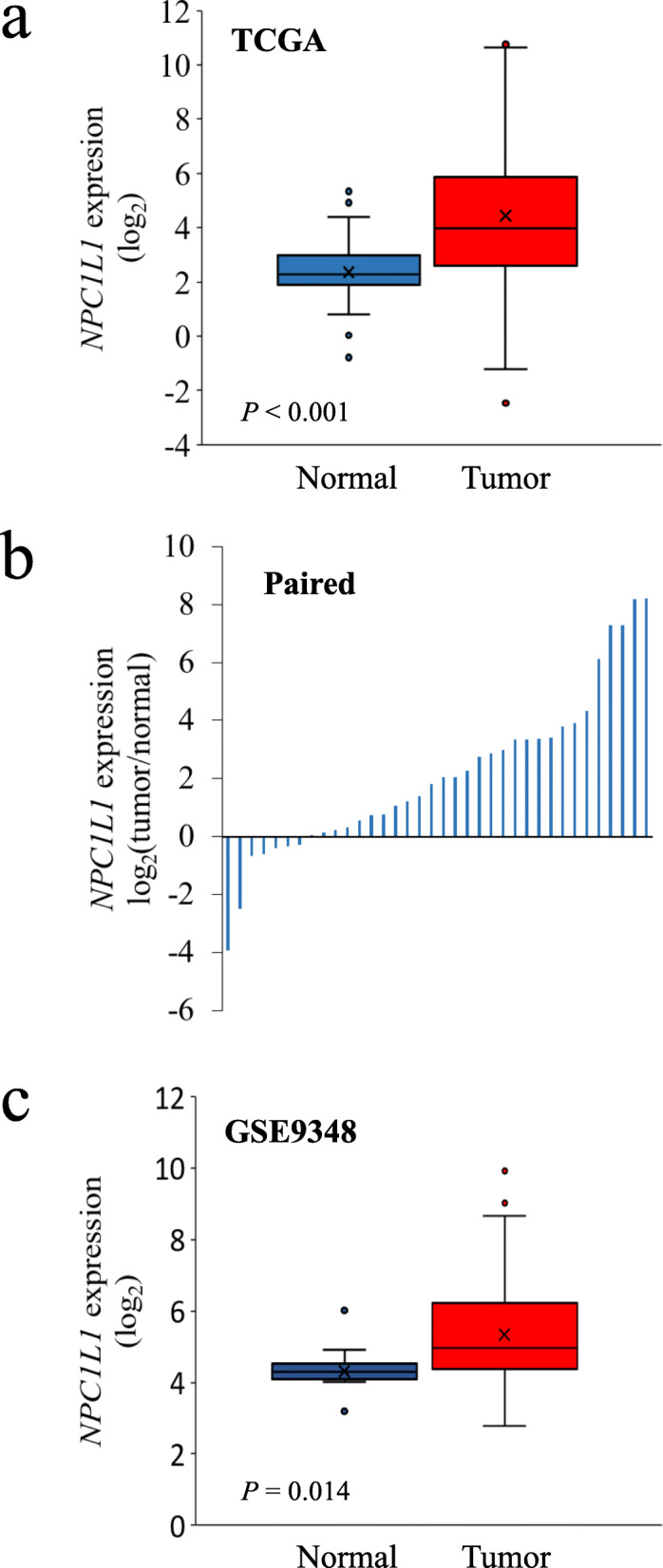
NPC1L1 expression in normal and CRC tissues. a NPC1L1 expression in CRC tissues as compared to that in normal tissues is presented as a box and whisker plot format. The numbers of normal and tumor tissues analyzed were 50 and 440, respectively. The mean value of NPC1L1 expression in normal tissues (blue box) was 7.00 and that in CRC tissues (red box) was 130.09. Significance was evaluated using the Mann–Whitney U test (P < 0.001). b Relative expression levels of NPC1L1 in tumor tissues (CRC) as compared to those in paired normal tissues are presented as a bar graph. The numbers of tumor and paired normal tissues analyzed were 35 (each). c NPC1L1 expression in CRC tissues as compared to that in normal tissues (GSE9348) is presented as a box and whisker plot format. The numbers of normal and tumor tissues analyzed were 12 and 70, respectively. The mean value of NPC1L1 expression in normal tissues (blue box) was 22.69 and that in CRC tissues (red box) was 81.35. Significance was evaluated using the Mann–Whitney U test (P = 0.014).
Relationship between NPC1L1 expression and patient characteristics
Based on the median value of NPC1L1 expression, the samples were divided into NPC1L1 low and NPC1L1 high expression groups and the correlation between NPC1L1 expression and the patient characteristics was examined (Table 2). The expression level of NPC1L1 was significantly correlated with the N stage (P < 0.05) and pathological stages (P < 0.05). However, no correlation was observed between NPC1L1 expression and the histological type, age, sex, and T and M stages.
Table 2.
Correlation between NPC1L1 expression and clinical characteristics of patients with CRC
| NPC1L1 expression | ||||
|---|---|---|---|---|
| Characteristic | N | Low | High | P-value |
| Histologic type | 423 | 0.257 | ||
| Adenocarcinoma | 186 | 177 | ||
| Mucinous | 26 | 34 | ||
| Age (years) | 428 | 0.923 | ||
| < 70 | 115 | 116 | ||
| ≥ 70 | 99 | 98 | ||
| Sex | 428 | 0.245 | ||
| Male | 120 | 108 | ||
| Female | 94 | 106 | ||
| T stages | 428 | 0.141 | ||
| Tis, T1–T2 | 47 | 35 | ||
| T3–T4 | 167 | 179 | ||
| M stages | 382 | 0.069 | ||
| M0 | 167 | 154 | ||
| M1 | 24 | 37 | ||
| N stages | 428 | 0.006 | ||
| N0 | 139 | 111 | ||
| N1–N2 | 75 | 103 | ||
| AJCC stages | 428 | 0.025 | ||
| Stage I–II | 132 | 109 | ||
| Stage III–IV | 82 | 105 | ||
AJCC American Joint Committee on Cancer, CRC colorectal cancer, NPC1L1 Niemann-Pick C1-Like 1
Five-year OS in the NPC1L1 low and NPC1L1 high expression groups
To determine whether NPC1L1 expression affects the prognosis of CRC patients, KM analysis was performed for the NPC1L1 low and NPC1L1 high expression groups against OS (NPC1L1 low expression group, n = 214; NPC1L1 high expression group, n = 214) (Fig. 3a). Based on the 75th percentile, the OS of the NPC1L1 low and NPC1L1 high expression groups was estimated to be 4.1 years and 2.3 years, respectively. Thus, the OS of CRC patients with high NPC1L1 expression was significantly less (P = 0.021) than that of CRC patients with low NPC1L1 expression. To confirm this result, CRC patients whose data were curated in two datasets, i.e., GSE17536 and GSE129451 were first stratified into NPC1L1 low and NPC1L1 high groups; 148 and 29 patients whose data were curated in the GSE17536 dataset were classified as NPC1L1 low and NPC1L1 high, respectively, whereas 48 and 14 patients whose data were curated in the GSE129451 dataset were classified as NPC1L1 low and NPC1L1 high, respectively. Consistent with the results obtained upon analyzing TCGA datasets, CRC patients classified as NPC1L1 high had significantly poorer survival than those classified as NPC1L1 low (P = 0.036 and 0.049) (Fig. 3b and c).
Fig. 3.
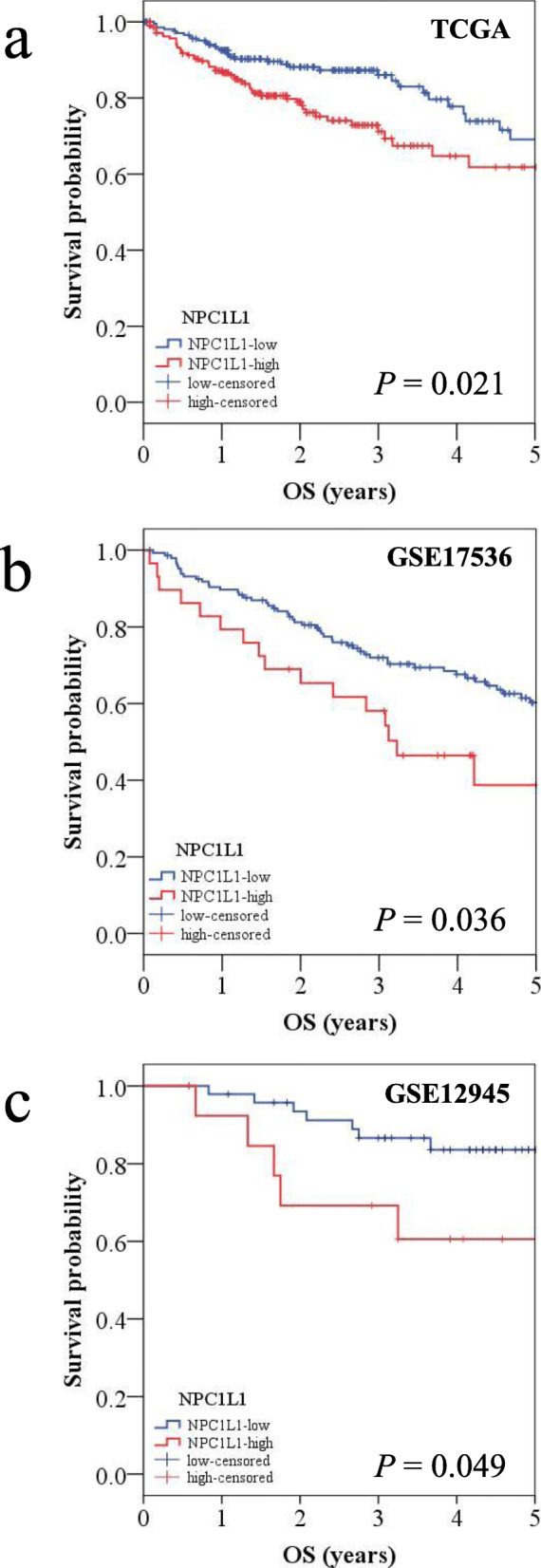
Kaplan–Meier survival analysis based on the expression level of NPC1L1. a Kaplan–Meier curve based on TCGA dataset. The blue line indicates the group with low NPC1L1 expression, and the red line indicates the group with high NPC1L1expression. The 75th percentiles of low and high NPC1L1 expression groups were 4.12 and 2.35 years, respectively. The P value was calculated using the log-rank method and was significant (P< 0.021). b, c Kaplan–Meier curve based on the GEO dataset (GSE17536 and GSE12945). The cutoff point was obtained using a ROC curve. The blue line indicates the group with low NPC1L1 expression, and the red line indicates the group with high NPC1L1 expression. The P value was calculated by the log-rank method and was significant (GSE17536, P = 0.036; GSE12945, P = 0.049)
Association between NPC1L1 expression and CRC prognosis
Univariate and multivariate Cox proportional hazards regression analyses were performed to assess the prognostic significance of NPC1L1 expression (Table 3). In the univariate analysis, age (P = 0.015), AJCC stage (P < 0.001), and NPC1L1 (P = 0.022) were identified as indicators of OS. Multivariate analysis revealed that age (P < 0.001), AJCC stage (P < 0.001), and NPC1L1 (P = 0.004) were significantly associated with OS. The hazard ratios for age, stage, and NPC1L1 were 2.286, 3.698, and 1.618, respectively. The prognostic importance of NPC1L1 with that of other markers, such as vascular endothelial growth factor A (VEGF-A), metastasis-associated in colon cancer 1 (MACC1), and transforming growth factor-beta 1 (TGF-β1) with respect to CRC was also compared (Table 4). Multivariate regression analysis revealed that the expression of NPC1L1 (P = 0.038) and VEGF-A (P = 0.009) was significantly associated with the OS of CRC patients.
Table 3.
Univariate and multivariate analyses to identify a correlation between prognostic factors and OS in CRC patients
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| HRa (95% CIb) | P-value | HRa (95% CIb) | P-value | |
| Age ≥ 70 (vs. < 70) | 1.669 (1.103–2.524) | 0.015 | 2.286 (1.485–3.519) | < 0.001 |
| Sex Male (vs. Female) | 0.872 (0.579–1.3174) | 0.513 | 0.942 (0.622–1.426) | 0.776 |
| Stage III + IV (vs. I + II) | 3.150 (2.044–4.855) | < 0.001 | 3.698 (2.366–5.779) | < 0.001 |
| NPC1L1 High (vs. Low) | 1.618 (1.071–2.445) | 0.022 | 1.618 (1.067–2.453) | 0.024 |
aHR hazard ratio, bCI confidence interval, OS overall survival, CRC colorectal cancer, NPC1L1 Niemann-Pick C1-Like 1
Table 4.
Univariate and multivariate analyses to identify a correlation between NPC1L1 expression and other prognostic factors of CRC
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| HRa (95% CIb) | P-value | HRa (95% CIb) | P-value | |
| NPC1L1 High (vs. Low) | 1.618 (1.071–2.445) | 0.022 | 1.557 (1.025–2.363) | 0.038 |
| VEGF-A High (vs. Low) | 1.796 (1.185–2.723) | 0.005 | 1.758 (1.150–2.686) | 0.009 |
| MACC1 High (vs. Low) | 1.567 (1.033–2.375) | 0.034 | 1.421 (0.928–2.178) | 0.106 |
| TGF-β1 High (vs. Low) | 1.369 (0.908–2.064) | 0.132 | 1.502 (0.990–2.278) | 0.056 |
aHR hazard ratio, bCI confidence interval, CRC colorectal cancer, NPC1L1 Niemann-Pick C1-Like 1, TGF-β1 transforming growth factor beta 1, MACC1 metastasis-associated in colon cancer 1, VEGF-A vascular endothelial growth factor-A
Survival curves after combining the prognostic factors
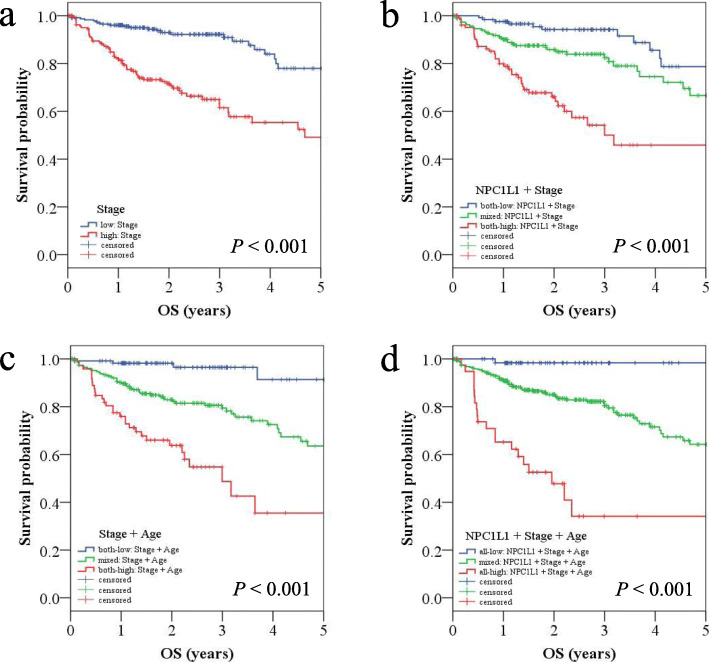
To identify CRC patients with a clinically worse prognosis, KM plots were generated using a combination of independent factors, such as stage, age, and NPC1L1 expression. Analysis of data corresponding to patients with CRC low and high stages revealed a median value of 4.68 years for patients with high stage CRC (Fig. 4a). Survival curves generated using a combination of two factors, i.e., NPC1L1 expression and stage revealed that the median value for CRC patients with the high stage/high NPC1L1 expression was 3.18 years, and that these patients showed a worse prognosis (Fig. 4b). Next, survival curves were generated using a combination of stage and age; these curves revealed the median value of 3.00 years for patients with high stage/high age, and they also revealed that these patients had a poor prognosis (Fig. 4c). Further, survival curves generated using a combination of three factors, i.e., stage, age, and NPC1L1 expression revealed that patients with high NPC1L1/high stage/high age had the shortest median value among all patients, i.e., 1.96 years (Fig. 4d).
Fig. 4.
Kaplan–Meier survival analysis after applying a combination of prognostic factors. The stage (low: I + II vs. high: III + IV), age (low: < 70 vs. high: ≥ 70), and NPC1L1 (low expression vs. high expression) groups were each divided into two groups. Then, survival analysis was performed by combining prognostic factors. The red line indicates a high stage and/or high NPC1L1 expression and/or older age group. The blue line indicates a low stage and/or low NPC1L1 expression and/or younger age group. The green line indicates mixed combination groups, excluding all high and all low combination groups. a Stage-only group. b NPC1L1 and stage group. c Stage and age group. d NPC1L1, stage, and age group
Discussion
NPC1L1 is essential for the intestinal absorption of cholesterol [10], and is a target of ezetimibe, a drug used to treat dyslipidemia recalcitrant to statin treatment. However, its expression and prognostic relevance in cancers have been poorly characterized. NPC1L1 expression was found to be downregulated in hepatocellular carcinoma (HCC) [14] but upregulated in pancreatic cancer [15]. In the present study, NPC1L1 expression was higher in CRC tissues than that in normal tissues (Fig. 2). A previous study revealed an inverse correlation between NPC1L1 expression and pathological stage, tumor differentiation, and vascular invasion in HCC [14]. In contrast, in the present study, NPC1L1 expression was positively associated with the N stage and pathological stage (Table 2). This can be explained by the fact that numerous studies have shown that the function of a specific gene in cancer development and progression may differ depending on the cancer type. For instance, increased Notch levels were correlated with tumor grade and metastasis in CRC [16], whereas Notch signaling was found to exhibit tumor-suppressive effects in glioblastoma [17]. Even in the same tumor cells, a gene may modulate cancer progression and suppression in an environment- or time-dependent manner [18, 19]. Thus, the involvement of NPC1L1 in cancer development may be cancer-type specific.
Abnormal lipid metabolism is associated with the development of vascular disorders, hyperlipidemia, lipid storage‒associated diseases, and obesity. Changes in lipid metabolism play an important role in the development and progression of cancer [20–22]. Lipids are used for energy supply, cell membrane structural material, signaling molecules, and post-transcriptional modification of cancer cells ; further, activation of lipogenic genes in cancer cells is known to promote cell proliferation and alter cell characteristics [20]. Studies have confirmed changes in the content and composition of fatty acids, oxylipins, and triacylglycerols in the serum, tumor tissue, and adipose tissue of CRC patients [21]. In addition, the molecules responsible for altered lipid metabolism are thought to serve as potential biomarkers for CRC development, progression, and prognosis [22]. Among them, NPC1L1 is known to be an important molecule for cholesterol absorption and is associated with patient prognosis in HCC [14]. The primary objective of this study was to investigate the difference in the 5-year survival between the NPC1L1 high and NPC1L1 low groups because this parameter is widely used to reflect the prognosis—and the endeavors required for the clinical management—of cancer. The results showed that patients in the NPC1L1 high expression group had poorer OS than those in the NPC1L1 low expression group (TCGA dataset) (Fig. 3a). Consistent with the results of this analysis, the analysis of datasets from other databases (GSE17536 and GSE129451) also revealed that high NPC1L1 expression was associated with poor survival outcomes in CRC patients (Fig. 3b and c). Univariate and multivariate analyses revealed that NPC1L1 expression, along with age and disease stage is significantly correlated with OS in CRC patients (Table 3). To confirm whether NPC1L1 represents an independent prognostic marker of CRC, its prognostic significance, along with that of other CRC prognostic markers, such as VEGF-A, MACC1, and TGF-β1, was determined using multivariate regression analysis (Table 4). Previous studies have shown that the OS of CRC patients with high expression of VEGF-A or MACC1 is significantly shorter than that of CRC patients with low expression of VEGF-A or MACC1 [23, 24]. A meta-analysis indicated the applicability of TGF-β as a prognostic factor for patients with CRC [25]. In this study, the expression of NPC1L1, along with that of VEGF-A, was significantly associated with CRC prognosis. Thus, these results suggest that NPC1L1, along with other known prognostic markers, is an independent prognostic marker for CRC.
Stage and age are recognized as prognostic factors related to mortality and survival in CRC [4]. To identify CRC patients with a worse prognosis in clinical practice based on NPC1L1 expression, KM plots generated using a combination of prognostic markers (NPC1L1, stage, and age) were used (Fig. 4). When a combination of NPC1L1 expression and stage was used to generate the survival curve, the median value for patients with high NPC1L1/high stage was lower than that of patients with high stage alone. Next, when a combination of NPC1L1 expression and stage/age was used to generate the survival curve, patients with high NPC1L1/high stage/high age exhibited a poorer prognosis than those with high stage/high age. Additionally, previous studies have shown that the apoptosis rate was higher in pancreatic cancer cells treated with ezetimibe or siNPC1L1 than that in control cells; this indicated an efficient method for treating pancreatic cancer [15]. These results suggest that NPC1L1 may be used as a screening marker to individually identify CRC patients with poor prognosis. Importantly, the findings of this study can serve as a basis for investigating the use of ezetimibe as an adjuvant treatment for CRC.
Strengths and limitations
The present study has several strengths. First, to our knowledge, this is the first study to reveal NPC1L1 as an independent prognostic factor in CRC. Thus, NPC1L1 may serve as a marker for enabling the clinical management of CRC patients, thereby improving the survival rates of these patients. Second, the datasets corresponding to data from a large number of CRC patients were used to demonstrate the relationship between high NPC1L1 expression and poor survival. Third, this study suggests an interesting future research direction for the employment of ezetimibe as an adjuvant treatment for CRC. Despite these strengths, the study has a few limitations. First, information regarding risk factors for CRC, such as family history, smoking habit, obesity, diabetes, and IBD, could not be presented owing to limited data. It may be interesting to identify the relationship between NPC1L1 expression and risk factors for CRC in the future. Second, no experimental verification for the development and progression of CRC resulting from high NPC1L1 expression was provided. In particular, the mRNA and protein expression does not always exhibit a proportional relationship. Depending on the gene in question, the mean correlation between mRNA and protein levels may vary from 0.28 to 0.71 [26]. Interestingly, the correlation between the mRNA and protein expression among cancer cell lines (osteosarcoma, epidermoid carcinoma, and glioblastoma) was high, i.e., from 0.58 to 0.63 [27]. Nicolle et al. found that the transcript-level expression of NPC1L1 was correlated with its protein level in pancreatic cancers [15]. If there is a high correlation between the mRNA and protein levels among cancer cells, the protein expression of NPC1L1 in CRC may also be high in proportion to the expressed NPC1L1 transcript. To confirm this expectation, further experimental studies are warranted. Although this study does not contribute any experimental evidence per se, it provides a framework for further studies aimed at experimentally investigating the role of NPC1L1 at the molecular and cellular levels in CRC.
Conclusions
This study shows, for the first time, that high NPC1L1 expression in CRC is associated with low survival. Thus, NPC1L1 can serve as an independent prognostic marker in CRC patients. Based on the results, clinicians will be able to use NPC1L1 expression as a marker to identify CRC patients with a poor prognosis. Moreover, the present study demonstrates NPC1L1 upregulation in CRC and its association with the N stage and pathological stages. These results will contribute toward our understanding of the relationship between CRC and cholesterol and experimentally investigating the role of NPC1L1 in the development and progression of CRC.
Acknowledgements
The authors would like to acknowledge the Cancer Genome Atlas (TCGA) database and the tools provided by TCGA database for data analysis.
Abbreviations
- AJCC
American Joint Committee on Cancer
- CI
Confidence interval
- CRC
Colorectal cancer
- GEO
Gene Expression Omnibus
- GDAC
Genome Data Analysis Center
- HCC
Hepatocellular carcinoma
- IBD
Inflammatory bowel disease
- KM
Kaplan-Meier
- MACC1
Metastasis-associated in colon cancer 1
- NPC1L1
Niemann-Pick C1-Like 1
- OS
Overall survival
- TCGA
The Cancer Genome Atlas
- TGF-β1
Transforming growth factor-beta 1
- VEGF-A
Vascular endothelial growth factor-A
Authors’ contributions
Conceptualization, R.J.K. and Y.H.C.; Formal analysis, R.J.K. and E.J.P.; Funding acquisition, Y.H.C.; Investigation, R.J.K., E.J.P. and Y.H.C.; Supervision, Y.H.C.; Validation, R.J.K., C.K., and C.H.; Writing – original draft, R.J.K. and Y.L.; Writing – review & editing, S.Y.L. All authors read and approved the final version of the manuscript.
Funding
This study was supported by the Research Institute for Convergence of Biomedical Science and Technology (30-2020-000), Pusan National University Yangsan Hospital.
Availability of data and materials
The datasets investigated and analyzed in this study are publicly available.
Declarations
Ethics approval and consent to participate
This manuscript does not include studies with human participants or animals; the authors have used the public datasets available in TCGA database.
Consent for publication
The authors provide formal consent to publish this work.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–78. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 4.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs RJ, Voorneveld PW, Kodach LL, Hardwick JC. Cholesterol metabolism and colorectal cancers. Curr Opin Pharmacol. 2012;12:690–5. doi: 10.1016/j.coph.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Nelson ER, Wardell SE, Jasper JS, Park S, Suchindran S, Howe MK, Carver NJ, Pillai RV, Sullivan PM, Sondhi V, et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science. 2013;342:1094–8. doi: 10.1126/science.1241908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelton K, Freeman MR, Solomon KR. Cholesterol and prostate cancer. Curr Opin Pharmacol. 2012;12:751–9. doi: 10.1016/j.coph.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarvinen R, Knekt P, Hakulinen T, Rissanen H, Heliovaara M. Dietary fat, cholesterol and colorectal cancer in a prospective study. Br J Cancer. 2001;85:357–61. doi: 10.1054/bjoc.2001.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stocks T, Lukanova A, Bjorge T, Ulmer H, Manjer J, Almquist M, Concin H, Engeland A, Hallmans G, Nagel G, et al. Metabolic factors and the risk of colorectal cancer in 580,000 men and women in the metabolic syndrome and cancer project (Me-Can) Cancer. 2011;117:2398–407. doi: 10.1002/cncr.25772. [DOI] [PubMed] [Google Scholar]
- 10.Betters JL, Yu L. NPC1L1 and cholesterol transport. FEBS Lett. 2010;584:2740–7. doi: 10.1016/j.febslet.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altmann SW, Davis HR, Jr, Zhu LJ, Yao X, Hoos LM, Tetzloff G, Iyer SP, Maguire M, Golovko A, Zeng M, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–4. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 12.He J, Shin H, Wei X, Kadegowda AK, Chen R, Xie SK. NPC1L1 knockout protects against colitis-associated tumorigenesis in mice. BMC Cancer. 2015;15:189. doi: 10.1186/s12885-015-1230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon RJ, Kim YH, Jeong DC, Han ME, Kim JY, Liu L, Jung JS, Oh SO. Expression and prognostic significance of zinc fingers and homeoboxes family members in renal cell carcinoma. PLoS One. 2017;12:e0171036. doi: 10.1371/journal.pone.0171036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen KJ, Jin RM, Shi CC, Ge RL, Hu L, Zou QF, Cai QY, Jin GZ, Wang K. The prognostic value of Niemann-Pick C1-like protein 1 and Niemann-Pick disease type C2 in hepatocellular carcinoma. J Cancer. 2018;9:556–63. doi: 10.7150/jca.19996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolle R, Blum Y, Marisa L, Loncle C, Gayet O, Moutardier V, Turrini O, Giovannini M, Bian B, Bigonnet M, et al. Pancreatic Adenocarcinoma Therapeutic Targets Revealed by Tumor-Stroma Cross-Talk Analyses in Patient-Derived Xenografts. Cell Rep. 2017;21:2458–70. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinson KE, George DC, Fender AW, Bertrand FE, Sigounas G. The Notch pathway in colorectal cancer. Int J Cancer. 2016;138:1835–42. doi: 10.1002/ijc.29800. [DOI] [PubMed] [Google Scholar]
- 17.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–51. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 18.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 19.Lebrun JJ. The Dual Role of TGFbeta in Human Cancer: From Tumor Suppression to Cancer Metastasis. ISRN Mol Biol. 2012;2012:381428. doi: 10.5402/2012/381428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: a review. Lipids Health Dis. 2019;18:29. doi: 10.1186/s12944-019-0977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long J, Zhang CJ, Zhu N, Du K, Yin YF, Tan X, Liao DF, Qin L. Lipid metabolism and carcinogenesis, cancer development. Am J Cancer Res. 2018;8:778–91. [PMC free article] [PubMed] [Google Scholar]
- 23.Jalba CS, Jalba BA, Nicula C, Zlatian O, Ioana M, Barca A, Cimpoeru A, Cruce M. Clinical relevance of vascular endothelial growth factor-A in colorectal cancer. Rom J Morphol Embryol. 2011;52:775–81. [PubMed] [Google Scholar]
- 24.Lin A, Zhang X, Zhang RL, He XF, Zhang JG, Yan WH. Prognostic and risk stratification value of lesion MACC1 expression in colorectal cancer patients. Front Oncol. 2019;9:28. doi: 10.3389/fonc.2019.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen XL, Chen ZQ, Zhu SL, Liu TW, Wen Y, Su YS, Xi XJ, Hu Y, Lian L, Liu FB. Prognostic value of transforming growth factor-beta in patients with colorectal cancer who undergo surgery: a meta-analysis. BMC Cancer. 2017;17:240. doi: 10.1186/s12885-017-3215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, Reimers MA, Scherf U, Kahn A, Dolginow D, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–32. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 27.Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, Algenäs C, Lundeberg J, Mann M, Uhlen M. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets investigated and analyzed in this study are publicly available.