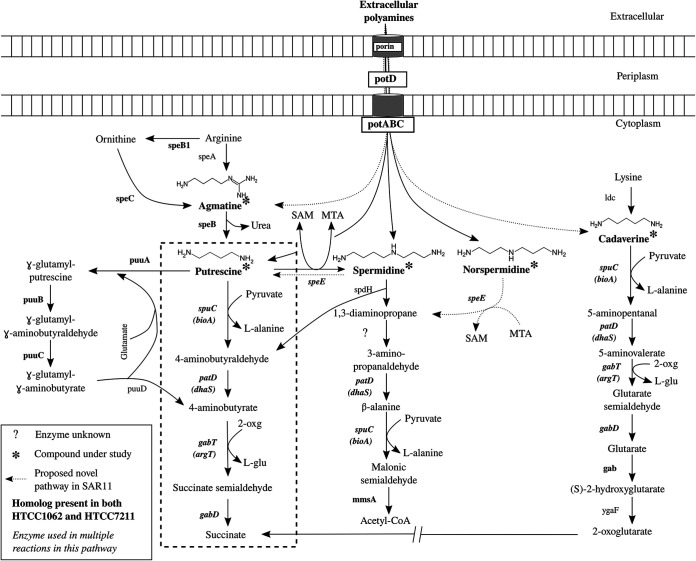
FIG 1.
Polyamine compound metabolism in SAR11. Common pathways for polyamine metabolism in bacteria are shown. The compounds under study are marked with asterisks, with enzymes listed in bold if both strains of SAR11 in this study, HTCC1062 and HTCC7211, have homologs to the enzyme, or in plain text if in neither. A question mark indicates that the enzyme is unknown. The enzyme name is in italics if it is used for multiple reactions in this metabolic system. The dashed box encompasses the pathway thought to be used by SAR11 cells for PUT metabolism based on previous studies. By-products of reactions where NH3 groups are transferred are included to show the flow of N. Gene names for SAR11 homologs, where different from canonical gene names are listed below the canonical name in parentheses. potC, spermidine/putrescine ABC transporter, permease; potB, permease; potD, SBP; potA, ATP-binding protein; speB1, arginase; speC, lysine/ornithine decarboxylase; speB, agmatinase; speE, spermidine/spermine synthase; puuA, gamma-glutamylputrescine synthetase; puuB, gamma-glutamylputrescine oxidoreductase; puuC, NADP/NAD-dependent aldehyde dehydrogenase; puuD, gamma-glutamyl-gamma-aminobutyrate hydrolase; spuC, putrescine-pyruvate aminotransferase; patD, 4-aminobutyraldehyde dehydrogenase; gabT, acetylornithine aminotransferase; gabD, succinate-semialdehyde dehydrogenase; spdH, spermidine dehydrogenase; mmsA, malonate-semialdehyde dehydrogenase; gab, glutarate 2-hydroxylase; ygaF, l-2-hydroxyglutarate dehydrogenase. SAM; S-adenosylmethionine; MTA, 5′-methylthioadenosine; 2-oxg, 2-oxoglutarate; l-glu, l-glutamate.