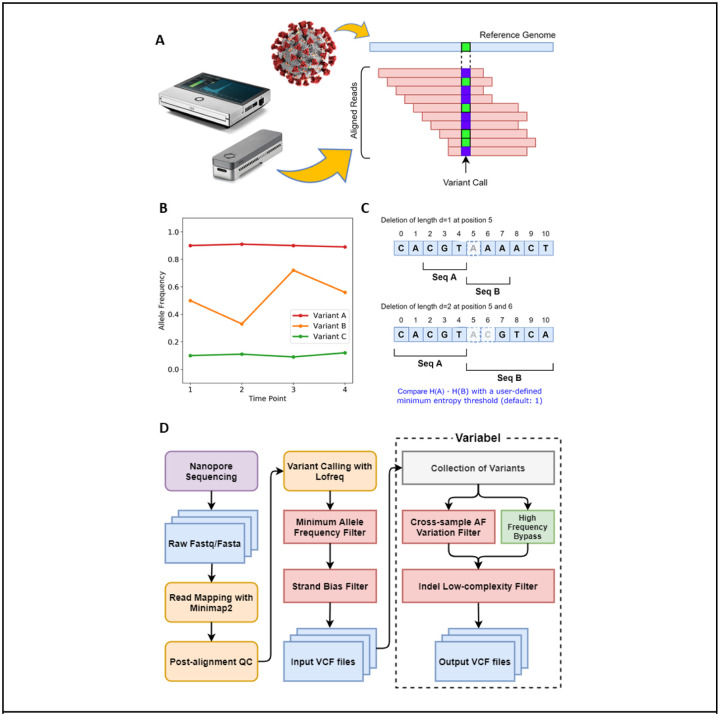
Figure 1. Illustration of Variabel algorithm and workflow.
a) Sequencing reads from ONT are aligned to the reference genome of SARS-CoV-2 with Minimap2, then variants are called based on the alignments using Lofreq. b) Cross-sample AF variantion filter identifies variants that are shared between samples. Variant calls with maximum AF less 0.65 and maximum AF variation less than 0.05 are classified as false calls. In this example, variant A and B pass the filter while variant C fails. c) Low-entropy filter calculates the Shannon’s entropy H for subsequences (Seq A and B) of the reference genome around the position where the indel call occurs. The length of the subsequences is determined by the length of the indel. Product of two entropy of the subsequences is used to determine whether the indel is a false positive or not. d) Workflow of Variabel for detecting intra-host variants for ONT sequences.